The Virtual Insect Brain protocol: creating and comparing standardized neuroanatomy
- PMID: 17196102
- PMCID: PMC1769402
- DOI: 10.1186/1471-2105-7-544
The Virtual Insect Brain protocol: creating and comparing standardized neuroanatomy
Abstract
Background: In the fly Drosophila melanogaster, new genetic, physiological, molecular and behavioral techniques for the functional analysis of the brain are rapidly accumulating. These diverse investigations on the function of the insect brain use gene expression patterns that can be visualized and provide the means for manipulating groups of neurons as a common ground. To take advantage of these patterns one needs to know their typical anatomy.
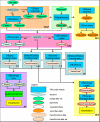
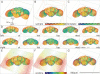
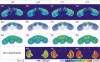
Results: This paper describes the Virtual Insect Brain (VIB) protocol, a script suite for the quantitative assessment, comparison, and presentation of neuroanatomical data. It is based on the 3D-reconstruction and visualization software Amira, version 3.x (Mercury Inc.) 1. Besides its backbone, a standardization procedure which aligns individual 3D images (series of virtual sections obtained by confocal microscopy) to a common coordinate system and computes average intensities for each voxel (volume pixel) the VIB protocol provides an elaborate data management system for data administration. The VIB protocol facilitates direct comparison of gene expression patterns and describes their interindividual variability. It provides volumetry of brain regions and helps to characterize the phenotypes of brain structure mutants. Using the VIB protocol does not require any programming skills since all operations are carried out at an intuitively usable graphical user interface. Although the VIB protocol has been developed for the standardization of Drosophila neuroanatomy, the program structure can be used for the standardization of other 3D structures as well.
Conclusion: Standardizing brains and gene expression patterns is a new approach to biological shape and its variability. The VIB protocol provides a first set of tools supporting this endeavor in Drosophila. The script suite is freely available at http://www.neurofly.de2.
Figures



References
-
- Amira http://www.amiravis.com
-
- project VIB. http://www.neurofly.de
-
- Brand AH, Perrimon N. Targeted gene expression as a means of altering cell fates and generating dominant phenotypes. Development. 1993;118:401–415. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

