Array-CGH and multipoint FISH to decode complex chromosomal rearrangements
- PMID: 17196103
- PMCID: PMC1769374
- DOI: 10.1186/1471-2164-7-330
Array-CGH and multipoint FISH to decode complex chromosomal rearrangements
Abstract
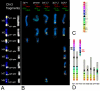
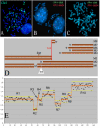
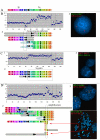
Background: Recently, several high-resolution methods of chromosome analysis have been developed. It is important to compare these methods and to select reliable combinations of techniques to analyze complex chromosomal rearrangements in tumours. In this study we have compared array-CGH (comparative genomic hybridization) and multipoint FISH (mpFISH) for their ability to characterize complex rearrangements on human chromosome 3 (chr3) in tumour cell lines. We have used 179 BAC/PAC clones covering chr3 with an approximately 1 Mb resolution to analyze nine carcinoma lines. Chr3 was chosen for analysis, because of its frequent rearrangements in human solid tumours.
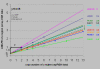
Results: The ploidy of the tumour cell lines ranged from near-diploid to near-pentaploid. Chr3 locus copy number was assessed by interphase and metaphase mpFISH. Totally 53 chr3 fragments were identified having copy numbers from 0 to 14. MpFISH results from the BAC/PAC clones and array-CGH gave mainly corresponding results. Each copy number change on the array profile could be related to a specific chromosome aberration detected by metaphase mpFISH. The analysis of the correlation between real copy number from mpFISH and the average normalized inter-locus fluorescence ratio (ANILFR) value detected by array-CGH demonstrated that copy number is a linear function of parameters that include the variable, ANILFR, and two constants, ploidy and background normalized fluorescence ratio.
Conclusion: In most cases, the changes in copy number seen on array-CGH profiles reflected cumulative chromosome rearrangements. Most of them stemmed from unbalanced translocations. Although our chr3 BAC/PAC array could identify single copy number changes even in pentaploid cells, mpFISH provided a more accurate analysis in the dissection of complex karyotypes at high ploidy levels.
Figures


Similar articles
-
Combined array-comparative genomic hybridization and single-nucleotide polymorphism-loss of heterozygosity analysis reveals complex genetic alterations in cervical cancer.BMC Genomics. 2007 Feb 20;8:53. doi: 10.1186/1471-2164-8-53. BMC Genomics. 2007. PMID: 17311676 Free PMC article.
-
High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays.Nat Genet. 1998 Oct;20(2):207-11. doi: 10.1038/2524. Nat Genet. 1998. PMID: 9771718
-
Effects of degenerate oligonucleotide-primed polymerase chain reaction amplification and labeling methods on the sensitivity and specificity of metaphase- and array-based comparative genomic hybridization.Cancer Genet Cytogenet. 2005 Apr 15;158(2):156-66. doi: 10.1016/j.cancergencyto.2004.08.033. Cancer Genet Cytogenet. 2005. PMID: 15796963
-
Molecular cytogenetic characterization of pancreas cancer cell lines reveals high complexity chromosomal alterations.Cytogenet Genome Res. 2007;118(2-4):148-56. doi: 10.1159/000108295. Cytogenet Genome Res. 2007. PMID: 18000365 Review.
-
[Analysis of genomic copy number alterations of malignant lymphomas and its application for diagnosis].Gan To Kagaku Ryoho. 2007 Jul;34(7):975-82. Gan To Kagaku Ryoho. 2007. PMID: 17637530 Review. Japanese.
Cited by
-
Segmental duplications and evolutionary plasticity at tumor chromosome break-prone regions.Genome Res. 2008 Mar;18(3):370-9. doi: 10.1101/gr.7010208. Epub 2008 Jan 29. Genome Res. 2008. PMID: 18230801 Free PMC article.
-
BAC CGH-array identified specific small-scale genomic imbalances in diploid DMBA-induced rat mammary tumors.BMC Cancer. 2012 Aug 15;12:352. doi: 10.1186/1471-2407-12-352. BMC Cancer. 2012. PMID: 22894538 Free PMC article.
-
Characterising chromosome rearrangements: recent technical advances in molecular cytogenetics.Heredity (Edinb). 2012 Jan;108(1):75-85. doi: 10.1038/hdy.2011.100. Epub 2011 Nov 16. Heredity (Edinb). 2012. PMID: 22086080 Free PMC article. Review.
-
Mandatory chromosomal segment balance in aneuploid tumor cells.BMC Cancer. 2007 Jan 26;7:21. doi: 10.1186/1471-2407-7-21. BMC Cancer. 2007. PMID: 17257397 Free PMC article.
-
Microenvironment-Dependent Phenotypic Changes in a SCID Mouse Model for Malignant Mesothelioma.Front Oncol. 2013 Aug 9;3:203. doi: 10.3389/fonc.2013.00203. eCollection 2013. Front Oncol. 2013. PMID: 23951555 Free PMC article.
References
-
- Van Prooijen-Knegt AC, Van Hoek JF, Bauman JG, Van Duijn P, Wool IG, Van der Ploeg M. In situ hybridization of DNA sequences in human metaphase chromosomes visualized by an indirect fluorescent immunocytochemical procedure. Exp Cell Res. 1982;141:397–407. doi: 10.1016/0014-4827(82)90228-2. - DOI - PubMed
-
- Pinkel D, Segraves R, Sudar D, Clark S, Poole I, Kowbel D, Collins C, Kuo WL, Chen C, Zhai Y, Dairkee SH, Ljung BM, Gray JW, Albertson DG. High resolution analysis of DNA copy number variation using comparative genomic hybridization to microarrays. Nat Genet. 1998;20:207–211. doi: 10.1038/2524. - DOI - PubMed
-
- Furuya T, Uchiyama T, Adachi A, Chochi Y, Oga A, Kawauchi S, Ishiglo K, Sasaki K. Relation of DNA ploidy to genetic aberrations detected by chromosomal CGH and FISH in gastric adenocarcinomas. Oncol Rep. 2006;15:1491–1496. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Miscellaneous

