Local encoding of computationally designed enzyme activity
- PMID: 17196220
- PMCID: PMC2963085
- DOI: 10.1016/j.jmb.2006.12.002
Local encoding of computationally designed enzyme activity
Abstract
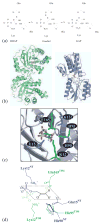
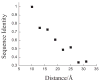
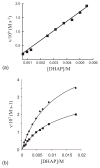
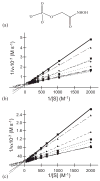
One aim of computational protein design is to introduce novel enzyme activity into proteins of known structure by predicting mutations that stabilize transition states. Previously, we showed that it is possible to introduce triose phosphate isomerase activity into the ribose-binding protein of Escherichia coli by constructing 17 mutations in the first two layers of residues that surround the wild-type ligand-binding site. Here, we report that these mutations can be "transplanted" into a homologous ribose-binding protein, isolated from the hyperthermophilic bacterium Thermoanaerobacter tengcongensis, with retention of catalytic activity, substrate affinity, and reaction pH dependence. The observed 10(5)-10(6)-fold rate enhancement corresponds to 70% of the maximally known transition-state binding energy. The wild-type sequences in these two homologues are almost perfectly conserved in the vicinity of their ribose-binding sites, but diverge significantly at increasing distance from these sites. The results demonstrate that the computationally designed mutations are sufficient to encode the observed enzyme activity, that all the observed activity is encoded locally within the layer of residues directly in contact with the substrate and that, in this case, at least 70% of transition state stabilization energy can be achieved using straightforward considerations of stereochemical complementarity between enzyme and reactants.
Figures

Similar articles
-
Computational design of a biologically active enzyme.Science. 2004 Jun 25;304(5679):1967-71. doi: 10.1126/science.1098432. Science. 2004. Retraction in: Science. 2008 Feb 1;319(5863):569. doi: 10.1126/science.319.5863.569b. PMID: 15218149 Retracted.
-
Biochemistry. De novo design of an enzyme.Science. 2004 Jun 25;304(5679):1916-7. doi: 10.1126/science.1100482. Science. 2004. PMID: 15218133 No abstract available.
-
Active site properties of monomeric triosephosphate isomerase (monoTIM) as deduced from mutational and structural studies.Protein Sci. 1996 Feb;5(2):229-39. doi: 10.1002/pro.5560050206. Protein Sci. 1996. PMID: 8745400 Free PMC article.
-
The backbone structure of the thermophilic Thermoanaerobacter tengcongensis ribose binding protein is essentially identical to its mesophilic E. coli homolog.BMC Struct Biol. 2008 Mar 28;8:20. doi: 10.1186/1472-6807-8-20. BMC Struct Biol. 2008. PMID: 18373848 Free PMC article.
-
The application of computational methods to the study of enzyme catalysis by triose-phosphate isomerase and stabilities of variants of bacteriophage T4 lysozyme.Ciba Found Symp. 1991;161:91-103; discussion 103-7. doi: 10.1002/9780470514146.ch7. Ciba Found Symp. 1991. PMID: 1814699 Review.
Cited by
-
Key protein-design papers challenged.Nature. 2009 Oct 15;461(7266):859. doi: 10.1038/461859a. Nature. 2009. PMID: 19829341 No abstract available.
References
-
- Wolfenden R, Snider MJ. The depth of chemical time and the power of enzymes as catalysts. Acc Chem Res. 2001;34:938–945. - PubMed
-
- Benkovic SJ, Hammes-Schiffer S. A perspective on enzyme catalysis. Science. 2003;301:1196–202. - PubMed
-
- Garcia-Viloca M, Gao J, Karplus M, Truhlar DG. How enzymes work: analysis by modern rate theory and computer simulations. Science. 2004;303:186–95. - PubMed
-
- Gutteridge A, Thornton JM. Understanding nature's catalytic toolkit. Trends Biochem Sci. 2005;30:622–9. - PubMed
-
- Torrance JW, Bartlett GJ, Porter CT, Thornton JM. Using a library of structural templates to recognise catalytic sites and explore their evolution in homologous families. J Mol Biol. 2005;347:565–81. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

