Multiple upstream modules regulate zebrafish myf5 expression
- PMID: 17199897
- PMCID: PMC1769357
- DOI: 10.1186/1471-213X-7-1
Multiple upstream modules regulate zebrafish myf5 expression
Abstract
Background: Myf5 is one member of the basic helix-loop-helix family of transcription factors, and it functions as a myogenic factor that is important for the specification and differentiation of muscle cells. The expression of myf5 is somite- and stage-dependent during embryogenesis through a delicate regulation. However, this complex regulatory mechanism of myf5 is not clearly understood.
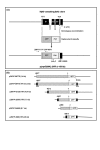
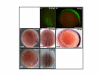
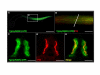
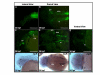
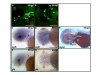
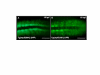
Results: We isolated a 156-kb bacterial artificial chromosome clone that includes an upstream 80-kb region and a downstream 70-kb region of zebrafish myf5 and generated a transgenic line carrying this 156-kb segment fused to a green fluorescent protein (GFP) reporter gene. We find strong GFP expression in the most rostral somite and in the presomitic mesoderm during segmentation stages, similar to endogenous myf5 expression. Later, the GFP signals persist in caudal somites near the tail bud but are down-regulated in the older, rostral somites. During the pharyngula period, we detect GFP signals in pectoral fin buds, dorsal rostral myotomes, hypaxial myotomes, and inferior oblique and superior oblique muscles, a pattern that also corresponds well with endogenous myf5 transcripts. To characterize the specific upstream cis-elements that regulate this complex and dynamic expression pattern, we also generated several transgenic lines that harbor various lengths within the upstream 80-kb segment. We find that (1) the -80 kb/-9977 segment contains a fin and cranial muscle element and a notochord repressor; (2) the -9977/-6213 segment contains a strong repressive element that does not include the notochord-specific repressor; (3) the -6212/-2938 segment contains tissue-specific elements for bone and spinal cord; (4) the -2937/-291 segment contains an eye enhancer, and the -2937/-2457 segment is required for notochord and myocyte expression; and (5) the -290/-1 segment is responsible for basal transcription in somites and the presomitic mesoderm.
Conclusion: We suggest that the cell lineage-specific expression of myf5 is delicately orchestrated by multiple modules within the distal upstream region. This study provides an insight to understand the molecular control of myf5 and myogenesis in the zebrafish.
Figures



References
-
- Weinberg ES, Allende ML, Kelly CS, Abdelhamid A, Murakami T, Andermann P, Doerre OG, Gruwald DJ, Riggleman B. Developmental regulation of zebrafish MyoD in wild-type, no tail and spadetail embryos. Development. 1996;122:271–280. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

