Structural basis of integrin regulation and signaling
- PMID: 17201681
- PMCID: PMC1952532
- DOI: 10.1146/annurev.immunol.25.022106.141618
Structural basis of integrin regulation and signaling
Abstract
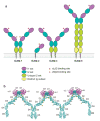
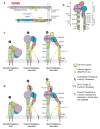
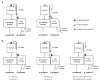
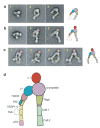
Integrins are cell adhesion molecules that mediate cell-cell, cell-extracellular matrix, and cell-pathogen interactions. They play critical roles for the immune system in leukocyte trafficking and migration, immunological synapse formation, costimulation, and phagocytosis. Integrin adhesiveness can be dynamically regulated through a process termed inside-out signaling. In addition, ligand binding transduces signals from the extracellular domain to the cytoplasm in the classical outside-in direction. Recent structural, biochemical, and biophysical studies have greatly advanced our understanding of the mechanisms of integrin bidirectional signaling across the plasma membrane. Large-scale reorientations of the ectodomain of up to 200 A couple to conformational change in ligand-binding sites and are linked to changes in alpha and beta subunit transmembrane domain association. In this review, we focus on integrin structure as it relates to affinity modulation, ligand binding, outside-in signaling, and cell surface distribution dynamics.
Figures
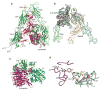
References
-
- Pribila JT, Quale AC, Mueller KL, Shimizu Y. Integrins and T cell-mediated immunity. Annu Rev Immunol. 2004;22:157–80. - PubMed
-
- Kinashi T. Intracellular signaling controlling integrin activation in lymphocytes. Nat Rev Immunol. 2005;5:546–59. - PubMed
-
- Anderson DC, Springer TA. Leukocyte adhesion deficiency: an inherited defect in the Mac-1, LFA-1, and p150,95 glycoproteins. Annu Rev Med. 1987;38:175–94. - PubMed
-
- Lee JO, Rieu P, Arnaout MA, Liddington R. Crystal structure of the A domain from the α subunit of integrin CR3 (CD11b/CD18) Cell. 1995;80:631–38. - PubMed
-
- Emsley J, Knight CG, Farndale RW, Barnes MJ, Liddington RC. Structural basis of collagen recognition by integrin α2β1. Cell. 2000;101:47–56. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

