Yeast genomic expression patterns in response to low-shear modeled microgravity
- PMID: 17201921
- PMCID: PMC1774566
- DOI: 10.1186/1471-2164-8-3
Yeast genomic expression patterns in response to low-shear modeled microgravity
Abstract
The low-shear microgravity environment, modeled by rotating suspension culture bioreactors called high aspect ratio vessels (HARVs), allows investigation in ground-based studies of the effects of microgravity on eukaryotic cells and provides insights into the impact of space flight on cellular physiology. We have previously demonstrated that low-shear modeled microgravity (LSMMG) causes significant phenotypic changes of a select group of Saccharomyces cerevisiae genes associated with the establishment of cell polarity, bipolar budding, and cell separation. However, the mechanisms cells utilize to sense and respond to microgravity and the fundamental gene expression changes that occur are largely unknown. In this study, we examined the global transcriptional response of yeast cells grown under LSMMG conditions using DNA microarray analysis in order to determine if exposure to LSMMG results in changes in gene expression.
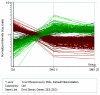
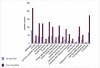
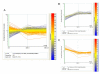
Results: LSMMG differentially changed the expression of a significant number of genes (1372) when yeast cells were cultured for either five generations or twenty-five generations in HARVs, as compared to cells grown under identical conditions in normal gravity. We identified genes in cell wall integrity signaling pathways containing MAP kinase cascades that may provide clues to novel physiological responses of eukaryotic cells to the external stress of a low-shear modeled microgravity environment. A comparison of the microgravity response to other environmental stress response (ESR) genes showed that 26% of the genes that respond significantly to LSMMG are involved in a general environmental stress response, while 74% of the genes may represent a unique transcriptional response to microgravity. In addition, we found changes in genes involved in budding, cell polarity establishment, and cell separation that validate our hypothesis that phenotypic changes observed in cells grown in microgravity are reflected in genome-wide changes. This study documents a considerable response to yeast cell growth in low-shear modeled microgravity that is evident, at least in part, by changes in gene expression. Notably, we identified genes that are involved in cell signaling pathways that allow cells to detect environmental changes, to respond within the cell, and to change accordingly, as well as genes of unknown function that may have a unique transcriptional response to microgravity. We also uncovered significant changes in the expression of many genes involved in cell polarization and bud formation that correlate well with the phenotypic effects observed in yeast cells when grown under similar conditions. These results are noteworthy as they have implications for human space flight.
Figures

Similar articles
-
Effects of low-shear modeled microgravity on cell function, gene expression, and phenotype in Saccharomyces cerevisiae.Appl Environ Microbiol. 2006 Jul;72(7):4569-75. doi: 10.1128/AEM.03050-05. Appl Environ Microbiol. 2006. PMID: 16820445 Free PMC article.
-
Response of Pseudomonas aeruginosa PAO1 to low shear modelled microgravity involves AlgU regulation.Environ Microbiol. 2010 Jun;12(6):1545-64. doi: 10.1111/j.1462-2920.2010.02184.x. Epub 2010 Mar 5. Environ Microbiol. 2010. PMID: 20236169
-
Gene expression and survival changes in Saccharomyces cerevisiae during suspension culture.Biotechnol Bioeng. 2006 Apr 20;93(6):1050-9. doi: 10.1002/bit.20810. Biotechnol Bioeng. 2006. PMID: 16440349
-
The genomics of yeast responses to environmental stress and starvation.Funct Integr Genomics. 2002 Sep;2(4-5):181-92. doi: 10.1007/s10142-002-0058-2. Epub 2002 Apr 30. Funct Integr Genomics. 2002. PMID: 12192591 Review.
-
Toward a genomic view of the gene expression program regulated by osmostress in yeast.OMICS. 2010 Dec;14(6):619-27. doi: 10.1089/omi.2010.0046. Epub 2010 Aug 20. OMICS. 2010. PMID: 20726780 Review.
Cited by
-
Bicluster Sampled Coherence Metric (BSCM) provides an accurate environmental context for phenotype predictions.BMC Syst Biol. 2015;9 Suppl 2(Suppl 2):S1. doi: 10.1186/1752-0509-9-S2-S1. Epub 2015 Apr 15. BMC Syst Biol. 2015. PMID: 25881257 Free PMC article.
-
Effects of Simulated Microgravity on the Proteome and Secretome of the Polyextremotolerant Black Fungus Knufia chersonesos.Front Genet. 2021 Mar 18;12:638708. doi: 10.3389/fgene.2021.638708. eCollection 2021. Front Genet. 2021. PMID: 33815472 Free PMC article.
-
The resolution of ambiguity as the basis for life: A cellular bridge between Western reductionism and Eastern holism.Prog Biophys Mol Biol. 2017 Dec;131:288-297. doi: 10.1016/j.pbiomolbio.2017.07.013. Epub 2017 Jul 22. Prog Biophys Mol Biol. 2017. PMID: 28743585 Free PMC article. Review.
-
Genomic Characterization of the Titan-like Cell Producing Naganishia tulchinskyi, the First Novel Eukaryote Isolated from the International Space Station.J Fungi (Basel). 2022 Feb 8;8(2):165. doi: 10.3390/jof8020165. J Fungi (Basel). 2022. PMID: 35205919 Free PMC article.
-
Growth and Antifungal Resistance of the Pathogenic Yeast, Candida Albicans, in the Microgravity Environment of the International Space Station: An Aggregate of Multiple Flight Experiences.Life (Basel). 2021 Mar 27;11(4):283. doi: 10.3390/life11040283. Life (Basel). 2021. PMID: 33801697 Free PMC article.
References
-
- Johanson K, Allen PL, Lewis F, Cubano LA, Hyman LE, Hammond TG. Saccharomyces cerevisiae gene expression changes during rotating wall vessel suspension culture. J Appl Physiol. 2002;93:2171–2180. - PubMed
-
- Johanson K, Allen PL, Gonzalez-Villalobos RA, Baker CB, D'Elia R, Hammond TG. Gene expression and survival changes in Saccharomyces cerevisiae during suspension culture. BiotechnolBioeng. 2006;93:1050–1059. - PubMed
-
- Brower CS, Sato S, Tomomori-Sato C, Kamura T, Pause A, Stearman R, Klausner RD, Malik S, Lane WS, Sorokina I, Roeder RG, Conaway JW, Conaway RC. Mammalian mediator subunit mMED8 is an Elongin BC-interacting protein that can assemble with Cul2 and Rbx1 to reconstitute a ubiquitin ligase. Proc Natl Acad Sci U S A. 2002;99:10353–10358. - PMC - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

