High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms
- PMID: 17202206
- PMCID: PMC1866008
- DOI: 10.1128/JVI.01643-06
High-frequency epigenetic repression and silencing of retroviruses can be antagonized by histone deacetylase inhibitors and transcriptional activators, but uniform reactivation in cell clones is restricted by additional mechanisms
Abstract
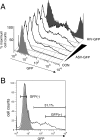
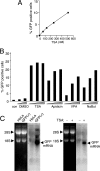
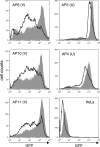
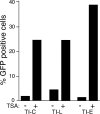
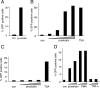
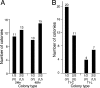
Integrated retroviral DNA is subject to epigenetic gene silencing, but the viral and host cell properties that influence initiation, maintenance, and reactivation are not fully understood. Here we describe rapid and high-frequency epigenetic repression and silencing of integrated avian sarcoma virus (ASV)-based vector DNAs in human HeLa cells. Initial studies utilized a vector carrying the strong human cytomegalovirus (hCMV) immediate-early (IE) promoter to drive expression of a green fluorescent protein (GFP) reporter gene, and cells were sorted into two populations based on GFP expression [GFP(+) and GFP(-)]. Two potent epigenetic effects were observed: (i) a very broad distribution of GFP intensities among cells in the GFP(+) population as well as individual GFP(+) clones and (ii) high-frequency GFP reporter gene silencing in GFP(-) cells. We previously showed that histone deacetylases (HDACs) can associate with ASV DNA soon after infection and may act to repress viral transcription at the level of chromatin. Consistent with this finding, we report here that treatment with the histone deacetylase inhibitor trichostatin A (TSA) induces GFP activation in GFP(-) cells and can also increase GFP expression in GFP(+) cells. In the case of the GFP(-) populations, we found that after removal of TSA, GFP silencing was reestablished in a subset of cells. We used that finding to enrich for stable GFP(-) cell populations in which viral GFP reporter expression could be reactivated by TSA; furthermore, we found that the ability to isolate such populations was independent of the promoter driving the GFP gene. In such enriched cultures, hCMV IE-driven, but not the viral long terminal repeat-driven, silent GFP reporter expression could be reactivated by the transcriptional activator prostratin. Microscopy-based studies using synchronized cells revealed variegated reactivation in cell clones, indicating that secondary epigenetic effects can restrict reactivation from silencing. Furthermore we found that entry into S phase was not required for reactivation. We conclude that HDACs can act rapidly to initiate and maintain promoter-independent retroviral epigenetic repression and silencing but that reactivation can be restricted by additional mechanisms.
Figures



References
-
- Baur, J. A., J. W. Shay, and W. E. Wright. 2004. Spontaneous reactivation of a silent telomeric transgene in a human cell line. Chromosoma 112:240-246. - PubMed
-
- Bushman, F., M. Lewinski, A. Ciuffi, S. Barr, J. Leipzig, S. Hannenhalli, and C. Hoffmann. 2005. Genome-wide analysis of retroviral DNA integration. Nat. Rev. Microbiol. 3:848-858. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

