Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6
- PMID: 17202207
- PMCID: PMC1865968
- DOI: 10.1128/JVI.02211-06
Efficient DNA packaging of bacteriophage PRD1 requires the unique vertex protein P6
Abstract
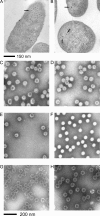
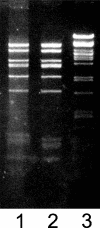
The assembly of bacteriophage PRD1 proceeds via formation of empty procapsids containing an internal lipid membrane, into which the linear double-stranded DNA genome is subsequently packaged. The packaging ATPase P9 and other putative packaging proteins have been shown to be located at a unique vertex of the PRD1 capsid. Here, we describe the isolation and characterization of a suppressor-sensitive PRD1 mutant deficient in the unique vertex protein P6. Protein P6 was found to be an essential part of the PRD1 packaging machinery; its absence leads to greatly reduced packaging efficiency. Lack of P6 was not found to affect particle assembly, because in the P6-deficient mutant infection, wild-type (wt) amounts of particles were produced, although most were empty. P6 was determined not to be a specificity factor, as the few filled particles seen in the P6-deficient infection contained only PRD1-specific DNA. The presence of P6 was not necessary for retention of DNA in the capsid once packaging had occurred, and P6-deficient DNA-containing particles were found to be stable and infectious, albeit not as infectious as wt PRD1 virions. A packaging model for bacteriophage PRD1, based on previous results and those obtained in this study, is presented.
Figures


Similar articles
-
In vitro DNA packaging of PRD1: a common mechanism for internal-membrane viruses.J Mol Biol. 2005 May 6;348(3):617-29. doi: 10.1016/j.jmb.2005.03.002. J Mol Biol. 2005. PMID: 15826659
-
The unique vertex of bacterial virus PRD1 is connected to the viral internal membrane.J Virol. 2003 Jun;77(11):6314-21. doi: 10.1128/jvi.77.11.6314-6321.2003. J Virol. 2003. PMID: 12743288 Free PMC article.
-
A structural model of the genome packaging process in a membrane-containing double stranded DNA virus.PLoS Biol. 2014 Dec 16;12(12):e1002024. doi: 10.1371/journal.pbio.1002024. eCollection 2014 Dec. PLoS Biol. 2014. PMID: 25514469 Free PMC article.
-
Lipid-containing viruses: bacteriophage PRD1 assembly.Adv Exp Med Biol. 2012;726:365-77. doi: 10.1007/978-1-4614-0980-9_16. Adv Exp Med Biol. 2012. PMID: 22297522 Review.
-
Membrane-Containing Icosahedral Bacteriophage PRD1: The Dawn of Viral Lineages.Adv Exp Med Biol. 2019;1215:85-109. doi: 10.1007/978-3-030-14741-9_5. Adv Exp Med Biol. 2019. PMID: 31317497 Review.
Cited by
-
Half a Century of Research on Membrane-Containing Bacteriophages: Bringing New Concepts to Modern Virology.Viruses. 2019 Jan 18;11(1):76. doi: 10.3390/v11010076. Viruses. 2019. PMID: 30669250 Free PMC article. Review.
-
Identification and characterization of a DNA binding domain on the adenovirus IVa2 protein.Virology. 2012 Nov 10;433(1):124-30. doi: 10.1016/j.virol.2012.07.013. Epub 2012 Aug 9. Virology. 2012. PMID: 22884292 Free PMC article.
-
Ternary complex formation on the adenovirus packaging sequence by the IVa2 and L4 22-kilodalton proteins.J Virol. 2007 Nov;81(22):12450-7. doi: 10.1128/JVI.01470-07. Epub 2007 Sep 5. J Virol. 2007. PMID: 17804492 Free PMC article.
-
Mechanism of membranous tunnelling nanotube formation in viral genome delivery.PLoS Biol. 2013 Sep;11(9):e1001667. doi: 10.1371/journal.pbio.1001667. Epub 2013 Sep 24. PLoS Biol. 2013. PMID: 24086111 Free PMC article.
-
Assembly and Evolution of Poxviruses.Adv Exp Med Biol. 2024;1451:35-54. doi: 10.1007/978-3-031-57165-7_3. Adv Exp Med Biol. 2024. PMID: 38801570 Review.
References
-
- Abrescia, N. G. A., J. J. B. Cockburn, J. M. Grimes, G. C. Sutton, J. M. Diprose, S. J. Butcher, S. D. Fuller, C. San Martin, R. M. Burnett, D. I. Stuart, D. H. Bamford, and J. K. H. Bamford. 2004. Insights into assembly from structural analysis of bacteriophage PRD1. Nature 432:68-74. - PubMed
-
- Agirrezabala, X., J. Martín-Benito, M. Valle, J. M. González, A. Valencia, J. M. Valpuesta, and J. L. Carrascosa. 2005. Structure of the connector of bacteriophage T7 at 8 Å resolution: structural homologies of a basic component of a DNA translocating machinery. J. Mol. Biol. 347:895-902. - PubMed
-
- Bamford, D. H., and H.-W. Ackermann. 2000. Family Tectiviridae, p. 111-116. In M. H. V. van Regenmortel, C. M. Fauquet, D. H. L. Bishop, E. B. Carstens, M. K. Estes, S. M. Lemon, J. Maniloff, M. A. Mayo, D. J. McGeoch, C. R. Pringle, and R. B. Wickner (ed.), Virus taxonomy: classification and nomenclature of viruses. Academic Press, San Diego, CA.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

