Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus
- PMID: 17202208
- PMCID: PMC1866046
- DOI: 10.1128/JVI.01939-06
Novel beta-barrel fold in the nuclear magnetic resonance structure of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus
Abstract
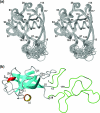
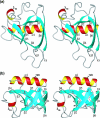
The nonstructural protein 1 (nsp1) of the severe acute respiratory syndrome coronavirus has 179 residues and is the N-terminal cleavage product of the viral replicase polyprotein that mediates RNA replication and processing. The specific function of nsp1 is not known. Here we report the nuclear magnetic resonance structure of the nsp1 segment from residue 13 to 128, which represents a novel alpha/beta-fold formed by a mixed parallel/antiparallel six-stranded beta-barrel, an alpha-helix covering one opening of the barrel, and a 3(10)-helix alongside the barrel. We further characterized the full-length 179-residue protein and show that the polypeptide segments of residues 1 to 12 and 129 to 179 are flexibly disordered. The structure is analyzed in a search for possible correlations with the recently reported activity of nsp1 in the degradation of mRNA.
Figures


Similar articles
-
Structure of alphacoronavirus transmissible gastroenteritis virus nsp1 has implications for coronavirus nsp1 function and evolution.J Virol. 2013 Mar;87(5):2949-55. doi: 10.1128/JVI.03163-12. Epub 2012 Dec 26. J Virol. 2013. PMID: 23269811 Free PMC article.
-
Identification of severe acute respiratory syndrome coronavirus replicase products and characterization of papain-like protease activity.J Virol. 2004 Dec;78(24):13600-12. doi: 10.1128/JVI.78.24.13600-13612.2004. J Virol. 2004. PMID: 15564471 Free PMC article.
-
Comprehensive structural analysis of designed incomplete polypeptide chains of the replicase nonstructural protein 1 from the severe acute respiratory syndrome coronavirus.PLoS One. 2017 Jul 27;12(7):e0182132. doi: 10.1371/journal.pone.0182132. eCollection 2017. PLoS One. 2017. PMID: 28750053 Free PMC article.
-
Hepatitis C virus RNA-dependent RNA polymerase (NS5B polymerase).Curr Top Microbiol Immunol. 2000;242:225-60. doi: 10.1007/978-3-642-59605-6_11. Curr Top Microbiol Immunol. 2000. PMID: 10592663 Review. No abstract available.
-
The PRRSV replicase: exploring the multifunctionality of an intriguing set of nonstructural proteins.Virus Res. 2010 Dec;154(1-2):61-76. doi: 10.1016/j.virusres.2010.07.030. Epub 2010 Aug 7. Virus Res. 2010. PMID: 20696193 Free PMC article. Review.
Cited by
-
The role of viral population diversity in adaptation of bovine coronavirus to new host environments.PLoS One. 2013;8(1):e52752. doi: 10.1371/journal.pone.0052752. Epub 2013 Jan 7. PLoS One. 2013. PMID: 23308119 Free PMC article.
-
Severe acute respiratory syndrome coronavirus protein nsp1 is a novel eukaryotic translation inhibitor that represses multiple steps of translation initiation.J Virol. 2012 Dec;86(24):13598-608. doi: 10.1128/JVI.01958-12. Epub 2012 Oct 3. J Virol. 2012. PMID: 23035226 Free PMC article.
-
In silico analysis of SARS-CoV-2 proteins as targets for clinically available drugs.Sci Rep. 2022 Mar 29;12(1):5320. doi: 10.1038/s41598-022-08320-y. Sci Rep. 2022. PMID: 35351926 Free PMC article.
-
Extended ensemble simulations of a SARS-CoV-2 nsp1-5'-UTR complex.PLoS Comput Biol. 2022 Jan 19;18(1):e1009804. doi: 10.1371/journal.pcbi.1009804. eCollection 2022 Jan. PLoS Comput Biol. 2022. PMID: 35045069 Free PMC article.
-
The Role of Viral RNA Degrading Factors in Shutoff of Host Gene Expression.Annu Rev Virol. 2022 Sep 29;9(1):213-238. doi: 10.1146/annurev-virology-100120-012345. Epub 2022 Jun 7. Annu Rev Virol. 2022. PMID: 35671567 Free PMC article. Review.
References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials

