ATP-dependent minor groove recognition of TA base pairs is required for template melting by the E1 initiator protein
- PMID: 17202221
- PMCID: PMC1866042
- DOI: 10.1128/JVI.02432-06
ATP-dependent minor groove recognition of TA base pairs is required for template melting by the E1 initiator protein
Abstract
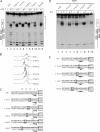
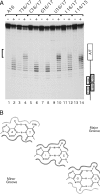
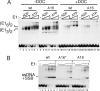
Template melting is an essential step in the initiation of DNA replication, but the mechanism of template melting is unknown for any replicon. Here we demonstrate that melting of the bovine papillomavirus type 1 ori is a sequence-dependent process which relies on specific recognition of TA base pairs in the minor groove by the E1 initiator. We show that correct template melting is a prerequisite for the formation of a stable double hexamer with helicase activity and that ori mutants that fail to melt correctly are defective for ori unwinding and DNA replication in vivo. Our results also indicate that melting of the DNA is achieved by destabilization of the double helix along its length through multiple interactions with E1, each of which is responsible for melting of a few base pairs, resulting in the extensive melting that is required for initiation of DNA replication.
Figures





Similar articles
-
Assembly of a double hexameric helicase.Mol Cell. 2005 Nov 11;20(3):377-89. doi: 10.1016/j.molcel.2005.09.020. Mol Cell. 2005. PMID: 16285920
-
A DNA-binding activity in BPV initiator protein E1 required for melting duplex ori DNA but not processive helicase activity initiated on partially single-stranded DNA.Nucleic Acids Res. 2008 Apr;36(6):1891-9. doi: 10.1093/nar/gkn041. Epub 2008 Feb 11. Nucleic Acids Res. 2008. PMID: 18267969 Free PMC article.
-
Adjacent residues in the E1 initiator beta-hairpin define different roles of the beta-hairpin in Ori melting, helicase loading, and helicase activity.Mol Cell. 2007 Mar 23;25(6):825-37. doi: 10.1016/j.molcel.2007.02.009. Mol Cell. 2007. PMID: 17386260
-
Transcription factor-dependent loading of the E1 initiator reveals modular assembly of the papillomavirus origin melting complex.J Biol Chem. 2000 Feb 4;275(5):3522-34. doi: 10.1074/jbc.275.5.3522. J Biol Chem. 2000. PMID: 10652347
-
DNA-directed base pair opening.Molecules. 2012 Oct 11;17(10):11947-64. doi: 10.3390/molecules171011947. Molecules. 2012. PMID: 23060287 Free PMC article. Review.
Cited by
-
Dynamic look at DNA unwinding by a replicative helicase.Proc Natl Acad Sci U S A. 2014 Mar 4;111(9):E827-35. doi: 10.1073/pnas.1322254111. Epub 2014 Feb 18. Proc Natl Acad Sci U S A. 2014. PMID: 24550505 Free PMC article.
-
A flexible brace maintains the assembly of a hexameric replicative helicase during DNA unwinding.Nucleic Acids Res. 2012 Mar;40(5):2271-83. doi: 10.1093/nar/gkr906. Epub 2011 Nov 8. Nucleic Acids Res. 2012. PMID: 22067453 Free PMC article.
-
Mechanistic analysis of local ori melting and helicase assembly by the papillomavirus E1 protein.Mol Cell. 2011 Sep 2;43(5):776-87. doi: 10.1016/j.molcel.2011.06.026. Mol Cell. 2011. PMID: 21884978 Free PMC article.
-
Origin remodeling and opening in bacteria rely on distinct assembly states of the DnaA initiator.J Biol Chem. 2010 Sep 3;285(36):28229-39. doi: 10.1074/jbc.M110.147975. Epub 2010 Jul 1. J Biol Chem. 2010. PMID: 20595381 Free PMC article.
-
Structure-based mutational analysis of the bovine papillomavirus E1 helicase domain identifies residues involved in the nonspecific DNA binding activity required for double trimer formation.J Virol. 2010 May;84(9):4264-76. doi: 10.1128/JVI.02214-09. Epub 2010 Feb 10. J Virol. 2010. PMID: 20147403 Free PMC article.
References
-
- Bell, S. P., and B. Stillman. 1992. ATP-dependent recognition of eukaryotic origins of DNA replication by a multiprotein complex. Nature 357:128-134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

