Positional clustering improves computational binding site detection and identifies novel cis-regulatory sites in mammalian GABAA receptor subunit genes
- PMID: 17204484
- PMCID: PMC1807961
- DOI: 10.1093/nar/gkl1062
Positional clustering improves computational binding site detection and identifies novel cis-regulatory sites in mammalian GABAA receptor subunit genes
Abstract
Understanding transcription factor (TF) mediated control of gene expression remains a major challenge at the interface of computational and experimental biology. Computational techniques predicting TF-binding site specificity are frequently unreliable. On the other hand, comprehensive experimental validation is difficult and time consuming. We introduce a simple strategy that dramatically improves robustness and accuracy of computational binding site prediction. First, we evaluate the rate of recurrence of computational TFBS predictions by commonly used sampling procedures. We find that the vast majority of results are biologically meaningless. However clustering results based on nucleotide position improves predictive power. Additionally, we find that positional clustering increases robustness to long or imperfectly selected input sequences. Positional clustering can also be used as a mechanism to integrate results from multiple sampling approaches for improvements in accuracy over each one alone. Finally, we predict and validate regulatory sequences partially responsible for transcriptional control of the mammalian type A gamma-aminobutyric acid receptor (GABA(A)R) subunit genes. Positional clustering is useful for improving computational binding site predictions, with potential application to improving our understanding of mammalian gene expression. In particular, predicted regulatory mechanisms in the mammalian GABA(A)R subunit gene family may open new avenues of research towards understanding this pharmacologically important neurotransmitter receptor system.
Figures



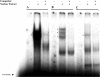

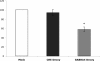
Similar articles
-
Integrating genomic data to predict transcription factor binding.Genome Inform. 2005;16(1):83-94. Genome Inform. 2005. PMID: 16362910
-
Identifying functional transcription factor binding sites in yeast by considering their positional preference in the promoters.PLoS One. 2013 Dec 26;8(12):e83791. doi: 10.1371/journal.pone.0083791. eCollection 2013. PLoS One. 2013. PMID: 24386279 Free PMC article.
-
Computational identification of transcription factor binding sites via a transcription-factor-centric clustering (TFCC) algorithm.J Mol Biol. 2002 Apr 19;318(1):71-81. doi: 10.1016/S0022-2836(02)00026-8. J Mol Biol. 2002. PMID: 12054769
-
A closer look at the high affinity benzodiazepine binding site on GABAA receptors.Curr Top Med Chem. 2011;11(2):241-6. doi: 10.2174/156802611794863562. Curr Top Med Chem. 2011. PMID: 21189125 Review.
-
GABAA receptors: building the bridge between subunit mRNAs, their promoters, and cognate transcription factors.Pharmacol Ther. 2004 Mar;101(3):259-81. doi: 10.1016/j.pharmthera.2003.12.002. Pharmacol Ther. 2004. PMID: 15031002 Review.
Cited by
-
Binding site graphs: a new graph theoretical framework for prediction of transcription factor binding sites.PLoS Comput Biol. 2007 May;3(5):e90. doi: 10.1371/journal.pcbi.0030090. Epub 2007 Apr 10. PLoS Comput Biol. 2007. PMID: 17500587 Free PMC article.
-
A computational approach for genome-wide mapping of splicing factor binding sites.Genome Biol. 2009;10(3):R30. doi: 10.1186/gb-2009-10-3-r30. Epub 2009 Mar 18. Genome Biol. 2009. PMID: 19296853 Free PMC article.
References
-
- Winderickx J., de Winde J.H., Crauwels M., Hino A., Hohmann S., Van Dijck P., Thevelein J.M. Regulation of genes encoding subunits of the trehalose synthase complex in Saccharomyces cerevisiae: novel variations of STRE-mediated transcription control? Mol. Gen. Genet. 1996;252:470–482. - PubMed
-
- Madhani H.D., Fink G.R. Combinatorial control required for the specificity of yeast MAPK signaling. Science. 1997;275:1314–1317. - PubMed
-
- Niehrs C., Pollet N. Synexpression groups in eukaryotes. Nature. 1999;402:483–487. - PubMed
-
- Lewin B. Genes VIII. Upper Saddle River, NJ: Pearson Prentice Hall; 2004.
-
- Stormo G.D. Consensus patterns in DNA. Methods Enzymol. 1990;183:211–221. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

