A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine
- PMID: 17204561
- PMCID: PMC1783357
- DOI: 10.1073/pnas.0610294104
A natural genetic code expansion cassette enables transmissible biosynthesis and genetic encoding of pyrrolysine
Abstract
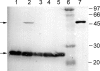
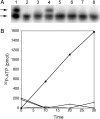
Pyrrolysine has entered natural genetic codes by the translation of UAG, a canonical stop codon. UAG translation as pyrrolysine requires the pylT gene product, an amber-decoding tRNA(Pyl) that is aminoacylated with pyrrolysine by the pyrrolysyl-tRNA synthetase produced from the pylS gene. The pylTS genes form a gene cluster with pylBCD, whose functions have not been investigated. The pylTSBCD gene order is maintained not only in methanogenic Archaea but also in a distantly related Gram-positive Bacterium, indicating past horizontal gene transfer of all five genes. Here we show that lateral transfer of pylTSBCD introduces biosynthesis and genetic encoding of pyrrolysine into a naïve organism. PylS-based assays demonstrated that pyrrolysine was biosynthesized in Escherichia coli expressing pylBCD from Methanosarcina acetivorans. Production of pyrrolysine did not require tRNA(Pyl) or PylS. However, when pylTSBCD were coexpressed with mtmB1, encoding the methanogen monomethylamine methyltransferase, UAG was translated as pyrrolysine to produce recombinant monomethylamine methyltransferase. Expression of pylTSBCD also suppressed an amber codon introduced into the E. coli uidA gene. Strains lacking one of the pylBCD genes did not produce pyrrolysine or translate UAG as pyrrolysine. These results indicated that pylBCD gene products biosynthesize pyrrolysine using metabolites common to Bacteria and Archaea and, furthermore, that the pyl gene cluster represents a "genetic code expansion cassette," previously unprecedented in natural organisms, whose transfer allows an existing codon to be translated as a novel endogenously synthesized free amino acid. Analogous cassettes may have served similar functions for other amino acids during the evolutionary expansion of the canonical genetic code.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
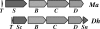

Similar articles
-
The complete biosynthesis of the genetically encoded amino acid pyrrolysine from lysine.Nature. 2011 Mar 31;471(7340):647-50. doi: 10.1038/nature09918. Nature. 2011. PMID: 21455182 Free PMC article.
-
In vivo contextual requirements for UAG translation as pyrrolysine.Mol Microbiol. 2007 Jan;63(1):229-41. doi: 10.1111/j.1365-2958.2006.05500.x. Epub 2006 Nov 27. Mol Microbiol. 2007. PMID: 17140411
-
Characterization of a Methanosarcina acetivorans mutant unable to translate UAG as pyrrolysine.Mol Microbiol. 2006 Jan;59(1):56-66. doi: 10.1111/j.1365-2958.2005.04927.x. Mol Microbiol. 2006. PMID: 16359318
-
Functional context, biosynthesis, and genetic encoding of pyrrolysine.Curr Opin Microbiol. 2011 Jun;14(3):342-9. doi: 10.1016/j.mib.2011.04.001. Epub 2011 May 5. Curr Opin Microbiol. 2011. PMID: 21550296 Free PMC article. Review.
-
tRNAPyl: Structure, function, and applications.RNA Biol. 2018;15(4-5):441-452. doi: 10.1080/15476286.2017.1356561. Epub 2017 Sep 13. RNA Biol. 2018. PMID: 28837402 Free PMC article. Review.
Cited by
-
Site-specific protein modifications through pyrroline-carboxy-lysine residues.Proc Natl Acad Sci U S A. 2011 Jun 28;108(26):10437-42. doi: 10.1073/pnas.1105197108. Epub 2011 Jun 13. Proc Natl Acad Sci U S A. 2011. PMID: 21670250 Free PMC article.
-
Pyrrolysyl-tRNA synthetase, an aminoacyl-tRNA synthetase for genetic code expansion.Croat Chem Acta. 2016 Jun;89(2):163-174. doi: 10.5562/cca2825. Epub 2016 Jun 14. Croat Chem Acta. 2016. PMID: 28239189 Free PMC article.
-
Engineering microbial hosts for production of bacterial natural products.Nat Prod Rep. 2016 Aug 27;33(8):963-87. doi: 10.1039/c6np00017g. Epub 2016 Apr 13. Nat Prod Rep. 2016. PMID: 27072804 Free PMC article. Review.
-
Translational recoding in archaea.Extremophiles. 2012 Nov;16(6):793-803. doi: 10.1007/s00792-012-0482-8. Epub 2012 Sep 27. Extremophiles. 2012. PMID: 23015064 Review.
-
Cellular Site-Specific Incorporation of Noncanonical Amino Acids in Synthetic Biology.Chem Rev. 2024 Sep 25;124(18):10577-10617. doi: 10.1021/acs.chemrev.3c00938. Epub 2024 Aug 29. Chem Rev. 2024. PMID: 39207844 Review.
References
-
- James CM, Ferguson TK, Leykam JF, Krzycki JA. J Biol Chem. 2001;276:34252–34258. - PubMed
-
- Hao B, Gong W, Ferguson TK, James CM, Krzycki JA, Chan MK. Science. 2002;296:1462–1466. - PubMed
-
- Hao B, Zhao G, Kang P, Soares J, Ferguson T, Gallucci J, Krzycki J, Chan M. Chem Biol. 2004;11:1317–1324. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

