Heterologous tissue culture expression signature predicts human breast cancer prognosis
- PMID: 17206280
- PMCID: PMC1764035
- DOI: 10.1371/journal.pone.0000145
Heterologous tissue culture expression signature predicts human breast cancer prognosis
Abstract
Background: Cancer patients have highly variable clinical outcomes owing to many factors, among which are genes that determine the likelihood of invasion and metastasis. This predisposition can be reflected in the gene expression pattern of the primary tumor, which may predict outcomes and guide the choice of treatment better than other clinical predictors.
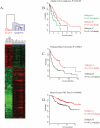
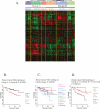
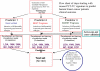
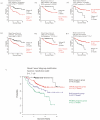
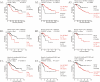
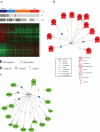
Methodology/principal findings: We developed an mRNA expression-based model that can predict prognosis/outcomes of human breast cancer patients regardless of microarray platform and patient group. Our model was developed using genes differentially expressed in mouse plasma cell tumors growing in vivo versus those growing in vitro. The prediction system was validated using published data from three cohorts of patients for whom microarray and clinical data had been compiled. The model stratified patients into four independent survival groups (BEST, GOOD, BAD, and WORST: log-rank test p = 1.7x10(-8)).
Conclusions: Our model significantly improved the survival prediction over other expression-based models and permitted recognition of patients with different prognoses within the estrogen receptor-positive group and within a single pathological tumor class. Basing our predictor on a dataset that originated in a different species and a different cell type may have rendered it less sensitive to proliferation differences and endowed it with wide applicability.
Significance: Prognosis prediction for patients with breast cancer is currently based on histopathological typing and estrogen receptor positivity. Yet both assays define groups that are heterogeneous in survival. Gene expression profiling allows subdivision of these groups and recognition of patients whose tumors are very unlikely to be lethal and those with much grimmer outlooks, which can augment the predictive power of conventional tumor analysis and aid the clinician in choosing relaxed vs. aggressive therapy.
Conflict of interest statement
Figures


References
-
- Kalluri R, Zeisberg M. Fibroblasts in cancer. Nature Reviews Cancer. 2006;6:392–401. - PubMed
-
- van de Vijver MJ, He YD, van't Veer LJ, Dai H, Hart AA, et al. A gene-expression signature as a predictor of survival in breast cancer. N Engl J Med. 2002;347:1999–2009. - PubMed
-
- van't Veer LJ, Dai H, van de Vijver MJ, He YD, Hart AA, et al. Gene expression profiling predicts clinical outcome of breast cancer. Nature. 2002;415:530–536. - PubMed
-
- Bild AH, Yao G, Chang JT, Wang Q, Potti A. Oncogenic pathway signatures in human cancers as a guide to targeted therapies. Nature. 2006;439:353–357. - PubMed
-
- Oh DS, Troester MA, Usary J, Hu Z, He X, et al. Estrogen-regulated genes predict survival in hormone receptor-positive breast cancers. J Clin Oncol. 2006;24:1656–1664. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials

