A high-resolution map of segmental DNA copy number variation in the mouse genome
- PMID: 17206864
- PMCID: PMC1761046
- DOI: 10.1371/journal.pgen.0030003
A high-resolution map of segmental DNA copy number variation in the mouse genome
Abstract
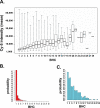
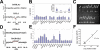
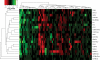
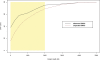
Submicroscopic (less than 2 Mb) segmental DNA copy number changes are a recently recognized source of genetic variability between individuals. The biological consequences of copy number variants (CNVs) are largely undefined. In some cases, CNVs that cause gene dosage effects have been implicated in phenotypic variation. CNVs have been detected in diverse species, including mice and humans. Published studies in mice have been limited by resolution and strain selection. We chose to study 21 well-characterized inbred mouse strains that are the focus of an international effort to measure, catalog, and disseminate phenotype data. We performed comparative genomic hybridization using long oligomer arrays to characterize CNVs in these strains. This technique increased the resolution of CNV detection by more than an order of magnitude over previous methodologies. The CNVs range in size from 21 to 2,002 kb. Clustering strains by CNV profile recapitulates aspects of the known ancestry of these strains. Most of the CNVs (77.5%) contain annotated genes, and many (47.5%) colocalize with previously mapped segmental duplications in the mouse genome. We demonstrate that this technique can identify copy number differences associated with known polymorphic traits. The phenotype of previously uncharacterized strains can be predicted based on their copy number at these loci. Annotation of CNVs in the mouse genome combined with sequence-based analysis provides an important resource that will help define the genetic basis of complex traits.
Conflict of interest statement
Competing interests. RRA, TAR, and PSE are employees of NimbleGen Systems, Inc.
Figures


References
-
- Sebat J, Lakshmi B, Troge J, Alexander J, Young J, et al. Large-scale copy number polymorphism in the human genome. Science. 2004;305:525–528. - PubMed
-
- Iafrate AJ, Feuk L, Rivera MN, Listewnik ML, Donahoe PK, et al. Detection of large-scale variation in the human genome. Nat Genet. 2004;36:949–951. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

