Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase
- PMID: 17207814
- PMCID: PMC1839928
- DOI: 10.1016/j.jmb.2006.12.013
Effects of DksA, GreA, and GreB on transcription initiation: insights into the mechanisms of factors that bind in the secondary channel of RNA polymerase
Abstract
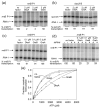
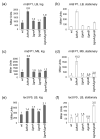
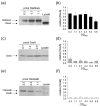
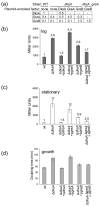
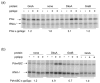
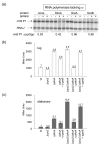
Escherichia coli DksA, GreA, and GreB have similar structures and bind to the same location on RNA polymerase (RNAP), the secondary channel. We show that GreB can fulfil some roles of DksA in vitro, including shifting the promoter-open complex equilibrium in the dissociation direction, thus allowing rRNA promoters to respond to changes in the concentration of ppGpp and NTPs. However, unlike deletion of the dksA gene, deletion of greB had no effect on rRNA promoters in vivo. We show that the apparent affinities of DksA and GreB for RNAP are similar, but the cellular concentration of GreB is much lower than that of DksA. When over-expressed and in the absence of competing GreA, GreB almost completely complemented the loss of dksA in control of rRNA expression, indicating its inability to regulate rRNA transcription in vivo results primarily from its low concentration. In contrast to GreB, the apparent affinity of GreA for RNAP was weaker than that of DksA, GreA affected rRNA promoters only modestly in vitro and, even when over-expressed, GreA did not affect rRNA transcription in vivo. Thus, binding in the secondary channel is necessary but insufficient to explain the effect of DksA on rRNA transcription. Neither Gre factor was capable of fulfilling two other functions of DksA in transcription initiation: co-activation of amino acid biosynthetic gene promoters with ppGpp and compensation for the loss of the omega subunit of RNAP in the response of rRNA promoters to ppGpp. Our results provide important clues to the mechanisms of both negative and positive control of transcription initiation by DksA.
Figures

References
-
- Record MT, Jr, Reznikoff WS, Craig ML, McQuade KL, Schlaz PJ. Escherichia coli RNA polymerase (Eσ70), promoters, and the kinetics of the steps of transcription initiation. In: Neidhardt FC, editor. Escherichia coli and Salmonella. ASM Press; Washington, D.C: 1996. pp. 792–820.
-
- Paul BJ, Ross W, Gaal T, Gourse RL. rRNA transcription in Escherichia coli. Annu Rev Genet. 2004;28:749–770. - PubMed
-
- Haugen SP, Berkman MB, Ross W, Gaal T, Ward C, Gourse RL. rRNA promoter regulation by nonoptimal binding of σ region 1.2: an additional recognition element for RNA polymerase. Cell. 2006;125:1069–1082. - PubMed
-
- Murray HD, Schneider DA, Gourse RL. Control of rRNA expression by small molecules is dynamic and nonredundant. Mol Cell. 2003;12:125–134. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

