hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes
- PMID: 17207965
- PMCID: PMC4647941
- DOI: 10.1016/j.ygeno.2006.11.012
hORFeome v3.1: a resource of human open reading frames representing over 10,000 human genes
Abstract
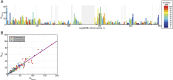
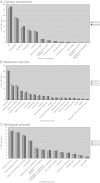
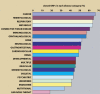
Complete sets of cloned protein-encoding open reading frames (ORFs), or ORFeomes, are essential tools for large-scale proteomics and systems biology studies. Here we describe human ORFeome version 3.1 (hORFeome v3.1), currently the largest publicly available resource of full-length human ORFs (available at ). Generated by Gateway recombinational cloning, this collection contains 12,212 ORFs, representing 10,214 human genes, and corresponds to a 51% expansion of the original hORFeome v1.1. An online human ORFeome database, hORFDB, was built and serves as the central repository for all cloned human ORFs (http://horfdb.dfci.harvard.edu). This expansion of the original ORFeome resource greatly increases the potential experimental search space for large-scale proteomics studies, which will lead to the generation of more comprehensive datasets.
Figures



References
-
- Adams M.D. The genome sequence of Drosophila melanogaster. Science. 2000;287:2185–2195. - PubMed
-
- Gibbs R.A. Genome sequence of the Brown Norway rat yields insights into mammalian evolution. Nature. 2004;428:493–521. - PubMed
-
- A. Goffeau, et al., Life with 6000 genes, Science 274 (1996) 546, 563–567. - PubMed
-
- Lander E.S. Initial sequencing and analysis of the human genome. Nature. 2001;409:860–921. - PubMed
-
- Venter J.C. The sequence of the human genome. Science. 2001;291:1304–1351. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

