A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria
- PMID: 17209024
- PMCID: PMC1899387
- DOI: 10.1128/JB.01491-06
A type IV-secretion-like system is required for conjugative DNA transport of broad-host-range plasmid pIP501 in gram-positive bacteria
Abstract
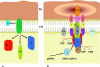
Plasmid pIP501 has a very broad host range for conjugative transfer among a wide variety of gram-positive bacteria and gram-negative Escherichia coli. Functionality of the pIP501 transfer (tra) genes in E. coli was proven by pIP501 retrotransfer to Enterococcus faecalis (B. Kurenbach, C. Bohn, J. Prabhu, M. Abudukerim, U. Szewzyk, and E. Grohmann, Plasmid 50:86-93, 2003). The 15 pIP501 tra genes are organized in a single operon (B. Kurenbach, J. Kopeć, M. Mägdefrau, K. Andreas, W. Keller, C. Bohn, M. Y. Abajy, and E. Grohmann, Microbiology 152:637-645, 2006). The pIP501 tra operon is negatively autoregulated at the transcriptional level by the conjugative DNA relaxase TraA. Three of the 15 pIP501-encoded Tra proteins show significant sequence similarity to the Agrobacterium type IV secretion system proteins VirB1, VirB4, and VirD4. Here we report a comprehensive protein-protein interaction map of all of the pIP501-encoded Tra proteins determined by the yeast two-hybrid assay. Most of the interactions were verified in vitro by isolation of the protein complexes with pull-down assays. In conjunction with known or postulated functions of the pIP501-encoded Tra proteins and computer-assisted prediction of their cellular location, we propose a model for the first type IV-secretion-like system encoded by a conjugative plasmid from gram-positive bacteria.
Figures
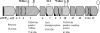



References
-
- Averhoff, B. 2004. DNA transport and natural transformation in mesophilic and thermophilic bacteria. J. Bioenerg. Biomembr. 36:25-33. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

