The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development
- PMID: 17209126
- PMCID: PMC1820972
- DOI: 10.1105/tpc.106.047506
The indeterminate gametophyte1 gene of maize encodes a LOB domain protein required for embryo Sac and leaf development
Abstract
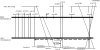
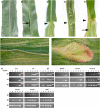
Angiosperm embryo sac development begins with a phase of free nuclear division followed by cellularization and differentiation of cell types. The indeterminate gametophyte1 (ig1) gene of maize (Zea mays) restricts the proliferative phase of female gametophyte development. ig1 mutant female gametophytes have a prolonged phase of free nuclear divisions leading to a variety of embryo sac abnormalities, including extra egg cells, extra polar nuclei, and extra synergids. Positional cloning of ig1 was performed based on the genome sequence of the orthologous region in rice. ig1 encodes a LATERAL ORGAN BOUNDARIES domain protein with high similarity to ASYMMETRIC LEAVES2 of Arabidopsis thaliana. A second mutant allele of ig1 was identified in a noncomplementation screen using active Mutator transposable element lines. Homozygous ig1 mutants have abnormal leaf morphology as well as abnormal embryo sac development. Affected leaves have disrupted abaxial-adaxial polarity and fail to repress the expression of meristem-specific knotted-like homeobox (knox) genes in leaf primordia, causing a proliferative, stem cell identity to persist in these cells. Despite the superficial similarity of ig1-O leaves and embryo sacs, ectopic knox gene expression cannot be detected in ig1-O embryo sacs.
Figures

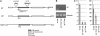






References
-
- Acharya, A., Ruvinov, S.B., Gal, J., Moll, J.R., and Vinson, C. (2002). A heterodimerizing leucine zipper coiled coil system for examining the specificity of a position interactions: amino acids I, V, L, N, A, and K. Biochemistry 41 14122–14131. - PubMed
-
- Banks, J.A. (1999). Gametophyte development in ferns. Annu. Rev. Plant Physiol. Plant Mol. Biol. 50 163–186. - PubMed
-
- Byrne, M.E., Barley, R., Curtis, M., Arroyo, J.M., Dunham, M., Hudson, A., and Martienssen, R.A. (2000). Asymmetric leaves1 mediates leaf patterning and stem cell function in Arabidopsis. Nature 408 967–971. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

