Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity
- PMID: 17209555
- PMCID: PMC2515318
- DOI: 10.1021/bi061243s
Human DNA polymerase alpha uses a combination of positive and negative selectivity to polymerize purine dNTPs with high fidelity
Abstract
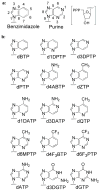
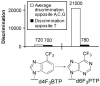
DNA polymerases accurately replicate DNA by incorporating mostly correct dNTPs opposite any given template base. We have identified the chemical features of purine dNTPs that human pol alpha uses to discriminate between right and wrong dNTPs. Removing N-3 from guanine and adenine, two high-fidelity bases, significantly lowers fidelity. Analogously, adding the equivalent of N-3 to low-fidelity benzimidazole-derived bases (i.e., bases that pol alpha rapidly incorporates opposite all four natural bases) and to generate 1-deazapurines significantly strengthens the ability of pol alpha to identify the resulting 1-deazapurines as wrong. Adding the equivalent of the purine N-1 to benzimidazole or to 1-deazapurines significantly decreases the rate at which pol alpha polymerizes the resulting bases opposite A, C, and G while simultaneously enhancing polymerization opposite T. Conversely, adding the equivalent of adenine's C-6 exocyclic amine (N-6) to 1- and 3-deazapurines also enhances polymerization opposite T but does not significantly decrease polymerization opposite A, C, and G. Importantly, if the newly inserted bases lack N-1 and N-6, pol alpha does not efficiently polymerize the next correct dNTP, whereas if it lacks N-3, one additional nucleotide is added and then chain termination ensues. These data indicate that pol alpha uses two orthogonal screens to maximize its fidelity. During dNTP polymerization, it uses a combination of negative (N-1 and N-3) and positive (N-1 and N-6) selectivity to differentiate between right and wrong dNTPs, while the shape of the base pair is essentially irrelevant. Then, to determine whether to add further dNTPs onto the just added nucleotide, pol alpha appears to monitor the shape of the base pair at the primer 3'-terminus. The biological implications of these results are discussed.
Figures






References
-
- Roberts JD, Kunkel TA. In: DNA Replication in Eukaryotic Cells: Concepts, Enzymes and Systems. Depamphilis M, editor. Cold Spring Harbor Laboratory Press; Cold Spring Harbor, NY: 1996. pp. 217–47.
-
- Kunkel TA, Bebenek K. Recent studies of the fidelity of DNA synthesis. Biochim Biophys Acta. 1988;951:1–5. - PubMed
-
- Watson JDCCFH. Molecular Structure of Nucleoic Acids. Nature. 1953;171:737–38. - PubMed
-
- Watson JD, Crick FHC. Genetical Implications of the Structure of Deoxyribonucleic Acid. Nature. 1953;171:964–67. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

