Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2'-OH group in regulating PutA--membrane binding
- PMID: 17209558
- PMCID: PMC2527739
- DOI: 10.1021/bi061935g
Redox-induced changes in flavin structure and roles of flavin N(5) and the ribityl 2'-OH group in regulating PutA--membrane binding
Abstract
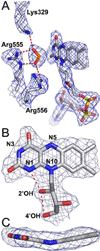
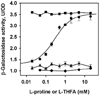
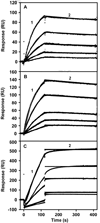
PutA is a novel flavoprotein in Escherichia coli that switches from a transcriptional repressor to a membrane-bound proline catabolic enzyme. Previous crystallographic studies of the PutA proline dehydrogenase (PRODH) domain under oxidizing conditions revealed that FAD N(5) and the ribityl 2'-OH group form hydrogen bonds with Arg431 and Arg556, respectively. Here we identify molecular interactions in the PutA PRODH active site that underlie redox-dependent functional switching of PutA. We report that reduction of the PRODH domain induces major structural changes in the FAD cofactor, including a 22 degrees bend of the isoalloxazine ring along the N(5)-N(10) axis, crankshaft rotation of the upper part of the ribityl chain, and formation of a new hydrogen bond network involving the ribityl 2'-OH group, FAD N(1), and Gly435. The roles of the FAD 2'-OH group and the FAD N(5)-Arg431 hydrogen bond pair in regulating redox-dependent PutA-membrane associations were tested using FAD analogues and site-directed mutagenesis. Kinetic membrane binding measurements and cell-based reporter gene assays of modified PutA proteins show that disrupting the FAD N(5)-Arg431 interaction impairs the reductive activation of PutA-membrane binding. We also show that the FAD 2'-OH group acts as a redox-sensitive toggle switch that controls PutA-membrane binding. These results illustrate a new versatility of the ribityl chain in flavoprotein mechanisms.
Figures



References
-
- Brown E, Wood JM. Redesigned purification yields a fully functional PutA protein dimer from Escherichia coli. J. Biol. Chem. 1992;267:13086–13092. - PubMed
-
- Menzel R, Roth J. Enzymatic properties of the purified putA protein from Salmonella typhimurium. J. Biol. Chem. 1981;256:9762–9766. - PubMed
-
- Menzel R, Roth J. Regulation of genes for proline utilization in Salmonella typhimurium: autogenous repression by the putA gene product. J. Mol. Biol. 1981;148:21–44. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

