Structural and functional analysis of the Na+/H+ exchanger
- PMID: 17209804
- PMCID: PMC1770851
- DOI: 10.1042/BJ20061062
Structural and functional analysis of the Na+/H+ exchanger
Abstract
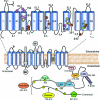
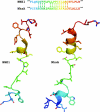
The mammalian NHE (Na+/H+ exchanger) is a ubiquitously expressed integral membrane protein that regulates intracellular pH by removing a proton in exchange for an extracellular sodium ion. Of the nine known isoforms of the mammalian NHEs, the first isoform discovered (NHE1) is the most thoroughly characterized. NHE1 is involved in numerous physiological processes in mammals, including regulation of intracellular pH, cell-volume control, cytoskeletal organization, heart disease and cancer. NHE comprises two domains: an N-terminal membrane domain that functions to transport ions, and a C-terminal cytoplasmic regulatory domain that regulates the activity and mediates cytoskeletal interactions. Although the exact mechanism of transport by NHE1 remains elusive, recent studies have identified amino acid residues that are important for NHE function. In addition, progress has been made regarding the elucidation of the structure of NHEs. Specifically, the structure of a single TM (transmembrane) segment from NHE1 has been solved, and the high-resolution structure of the bacterial Na+/H+ antiporter NhaA has recently been elucidated. In this review we discuss what is known about both functional and structural aspects of NHE1. We relate the known structural data for NHE1 to the NhaA structure, where TM IV of NHE1 shows surprising structural similarity with TM IV of NhaA, despite little primary sequence similarity. Further experiments that will be required to fully understand the mechanism of transport and regulation of the NHE1 protein are discussed.
Figures
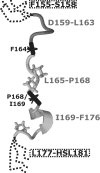
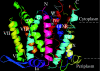

References
-
- Orlowski J., Grinstein S. Na+/H+ exchangers of mammalian cells. J. Biol. Chem. 1997;272:22373–22376. - PubMed
-
- Karmazyn M., Gan T., Humphreys R. A., Yoshida H., Kusumoto K. The myocardial Na+–H+ exchange. Structure, regulation, and its role in heart disease. Circ. Res. 1999;85:777–786. - PubMed
-
- Cardone R. A., Casavola V., Reshkin S. J. The role of disturbed pH dynamics and the Na+/H+ exchanger in metastasis. Nat. Rev. Cancer. 2005;5:786–795. - PubMed
-
- Slepkov E. R., Rainey J. K., Li X., Liu Y., Cheng F. J., Lindhout D. A., Sykes B. D., Fliegel L. Structural and functional characterization of transmembrane segment IV of the NHE1 isoform of the Na+/H+ exchanger. J. Biol. Chem. 2005;280:17863–17872. - PubMed
-
- Orlowski J., Grinstein S. Diversity of the mammalian sodium/proton exchanger SLC9 gene family. Pflügers Arch. 2004;447:549–565. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

