CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide
- PMID: 17210645
- PMCID: PMC1820452
- DOI: 10.1128/MCB.01993-06
CTCF interacts with and recruits the largest subunit of RNA polymerase II to CTCF target sites genome-wide
Abstract
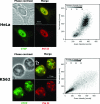
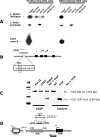
CTCF is a transcription factor with highly versatile functions ranging from gene activation and repression to the regulation of insulator function and imprinting. Although many of these functions rely on CTCF-DNA interactions, it is an emerging realization that CTCF-dependent molecular processes involve CTCF interactions with other proteins. In this study, we report the association of a subpopulation of CTCF with the RNA polymerase II (Pol II) protein complex. We identified the largest subunit of Pol II (LS Pol II) as a protein significantly colocalizing with CTCF in the nucleus and specifically interacting with CTCF in vivo and in vitro. The role of CTCF as a link between DNA and LS Pol II has been reinforced by the observation that the association of LS Pol II with CTCF target sites in vivo depends on intact CTCF binding sequences. "Serial" chromatin immunoprecipitation (ChIP) analysis revealed that both CTCF and LS Pol II were present at the beta-globin insulator in proliferating HD3 cells but not in differentiated globin synthesizing HD3 cells. Further, a single wild-type CTCF target site (N-Myc-CTCF), but not the mutant site deficient for CTCF binding, was sufficient to activate the transcription from the promoterless reporter gene in stably transfected cells. Finally, a ChIP-on-ChIP hybridization assay using microarrays of a library of CTCF target sites revealed that many intergenic CTCF target sequences interacted with both CTCF and LS Pol II. We discuss the possible implications of our observations with respect to plausible mechanisms of transcriptional regulation via a CTCF-mediated direct link of LS Pol II to the DNA.
Figures





References
-
- Acker, J., M. de Graaff, I. Cheynel, V. Khazak, C. Kedinger, and M. Vigneron. 1997. Interactions between the human RNA polymerase II subunits. J. Biol. Chem. 272:16815-16821. - PubMed
-
- Bell, A. C., and G. Felsenfeld. 2000. Methylation of a CTCF-dependent boundary controls imprinted expression of the Igf2 gene. Nature 405:482-485. - PubMed
-
- Bell, A. C., A. G. West, and G. Felsenfeld. 1999. The protein CTCF is required for the enhancer blocking activity of vertebrate insulators. Cell 98:387-396. - PubMed
-
- Beug, H., G. Doederlein, C. Freudenstein, and T. Graf. 1982. Erythroblast cell lines transformed by a temperature-sensitive mutant of avian erythroblastosis virus: a model system to study erythroid differentiation in vitro. J. Cell. Physiol. Suppl. 1:195-207. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
