Igkappa allelic inclusion is a consequence of receptor editing
- PMID: 17210730
- PMCID: PMC2118438
- DOI: 10.1084/jem.20061918
Igkappa allelic inclusion is a consequence of receptor editing
Abstract
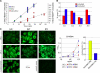
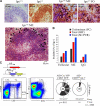
The discovery of lymphocytes bearing two light chains in mice carrying self-reactive antibody transgenes has challenged the "one lymphocyte-one antibody" rule. However, the extent and nature of allelically included cells in normal mice is unknown. We show that 10% of mature B cells coexpress both Igkappa alleles. These cells are not the result of failure in allelic exclusion per se, but arise through receptor editing. We find that under physiological conditions, editing occurs both by deletion and by inclusion with equal probability. In addition, we demonstrate that B lymphocytes carrying two B-cell receptors are recruited to germinal center reactions, and thus fully participate in humoral immune responses. Our data measure the scope of allelic inclusion and provide a mechanism whereby autoreactive B cells might "escape" central tolerance.
Figures


References
-
- Tonegawa, S. 1983. Somatic generation of antibody diversity. Nature. 302:575–581. - PubMed
-
- Goodnow, C.C., J. Crosbie, S. Adelstein, T.B. Lavoie, S.J. Smith-Gill, R.A. Brink, H. Pritchard-Briscoe, J.S. Wotherspoon, R.H. Loblay, K. Raphael, et al. 1988. Altered immunoglobulin expression and functional silencing of self-reactive B lymphocytes in transgenic mice. Nature. 334:676–682. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

