pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene
- PMID: 17210787
- PMCID: PMC1759899
- DOI: 10.1101/gad.1499407
pRB family proteins are required for H3K27 trimethylation and Polycomb repression complexes binding to and silencing p16INK4alpha tumor suppressor gene
Abstract
Genetic studies have demonstrated that Bmi1 promotes cell proliferation and stem cell self-renewal with a correlative decrease of p16(INK4a) expression. Here, we demonstrate that Polycomb genes EZH2 and BMI1 repress p16 expression in human and mouse primary cells, but not in cells deficient for pRB protein function. The p16 locus is H3K27-methylated and bound by BMI1, RING2, and SUZ12. Inactivation of pRB family proteins abolishes H3K27 methylation and disrupts BMI1, RING2, and SUZ12 binding to the p16 locus. These results suggest a model in which pRB proteins recruit PRC2 to trimethylate p16, priming the BMI1-containing PRC1L ubiquitin ligase complex to silence p16.
Figures



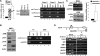
References
-
- Bruggeman S.W., Valk-Lingbeek M.E., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Valk-Lingbeek M.E., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., van der Stoop P.P., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Jacobs J.J., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Kieboom K., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Tanger E., Hulsman D., Leung C., Arsenijevic Y., Marino S., Hulsman D., Leung C., Arsenijevic Y., Marino S., Leung C., Arsenijevic Y., Marino S., Arsenijevic Y., Marino S., Marino S., et al. Ink4a and Arf differentially affect cell proliferation and neural stem cell self-renewal in Bmi1-deficient mice. Genes & Dev. 2005;19:1438–1443. - PMC - PubMed
-
- Cao R., Tsukada Y., Zhang Y., Tsukada Y., Zhang Y., Zhang Y. Role of Bmi-1 and Ring1A in H2A ubiquitylation and Hox gene silencing. Mol. Cell. 2005;20:845–854. - PubMed
-
- Cobrinik D. Pocket proteins and cell cycle control. Oncogene. 2005;24:2796–2809. - PubMed
-
- Hara E., Smith R., Parry D., Tahara H., Stone S., Peters G., Smith R., Parry D., Tahara H., Stone S., Peters G., Parry D., Tahara H., Stone S., Peters G., Tahara H., Stone S., Peters G., Stone S., Peters G., Peters G. Regulation of p16CDKN2 expression and its implication for cell immortalization and senescence. Mol. Cell. Biol. 1996;16:859–867. - PMC - PubMed
-
- Isono K., Fujimura Y., Shinga J., Yamaki M., O-Wang J., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Fujimura Y., Shinga J., Yamaki M., O-Wang J., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Shinga J., Yamaki M., O-Wang J., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Yamaki M., O-Wang J., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., O-Wang J., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Takihara Y., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Murahashi Y., Takada Y., Mizutani-Koseki Y., Koseki H., Takada Y., Mizutani-Koseki Y., Koseki H., Mizutani-Koseki Y., Koseki H., Koseki H. Mammalian polyhomeotic homologues Phc2 and Phc1 act in synergy to mediate Polycomb repression of Hox genes. Mol. Cell. Biol. 2005;25:6694–6706. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
