Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer
- PMID: 17210928
- PMCID: PMC1781351
- DOI: 10.1101/gr.5942807
Reductive evolution and niche adaptation inferred from the genome of Mycobacterium ulcerans, the causative agent of Buruli ulcer
Abstract
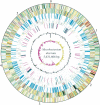
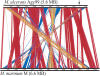
Mycobacterium ulcerans is found in aquatic ecosystems and causes Buruli ulcer in humans, a neglected but devastating necrotic disease of subcutaneous tissue that is rampant throughout West and Central Africa. Here, we report the complete 5.8-Mb genome sequence of M. ulcerans and show that it comprises two circular replicons, a chromosome of 5632 kb and a virulence plasmid of 174 kb. The plasmid is required for production of the polyketide toxin mycolactone, which provokes necrosis. Comparisons with the recently completed 6.6-Mb genome of Mycobacterium marinum revealed >98% nucleotide sequence identity and genome-wide synteny. However, as well as the plasmid, M. ulcerans has accumulated 213 copies of the insertion sequence IS2404, 91 copies of IS2606, 771 pseudogenes, two bacteriophages, and multiple DNA deletions and rearrangements. These data indicate that M. ulcerans has recently evolved via lateral gene transfer and reductive evolution from the generalist, more rapid-growing environmental species M. marinum to become a niche-adapted specialist. Predictions based on genome inspection for the production of modified mycobacterial virulence factors, such as the highly abundant phthiodiolone lipids, were confirmed by structural analyses. Similarly, 11 protein-coding sequences identified as M. ulcerans-specific by comparative genomics were verified as such by PCR screening a diverse collection of 33 strains of M. ulcerans and M. marinum. This work offers significant insight into the biology and evolution of mycobacterial pathogens and is an important component of international efforts to counter Buruli ulcer.
Figures


References
-
- Alsop D. The Bairnsdale ulcer. Aust. N. Z. J. Surg. 1972;41:317–319. - PubMed
-
- Brodin P., Rosenkrands I., Andersen P., Cole S.T., Brosch R., Rosenkrands I., Andersen P., Cole S.T., Brosch R., Andersen P., Cole S.T., Brosch R., Cole S.T., Brosch R., Brosch R. ESAT-6 proteins: Protective antigens and virulence factors? Trends Microbiol. 2004;12:500–508. - PubMed
-
- Carver T.J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rutherford K.M., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Berriman M., Rajandream M.A., Barrell B.G., Parkhill J., Rajandream M.A., Barrell B.G., Parkhill J., Barrell B.G., Parkhill J., Parkhill J. ACT: The Artemis Comparison Tool. Bioinformatics. 2005;21:3422–3423. - PubMed
-
- Cole S.T., Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Brosch R., Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Parkhill J., Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Garnier T., Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Churcher C., Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Harris D., Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Gordon S.V., Eiglmeier K., Gas S., Barry C.E., III, Eiglmeier K., Gas S., Barry C.E., III, Gas S., Barry C.E., III, Barry C.E., III, et al. Deciphering the biology of Mycobacterium tuberculosis from the complete genome sequence. Nature. 1998;393:537–544. - PubMed
-
- Cole S.T., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Eiglmeier K., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Parkhill J., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., James K.D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Thomson N.R., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Wheeler P.R., Honore N., Garnier T., Churcher C., Harris D., Honore N., Garnier T., Churcher C., Harris D., Garnier T., Churcher C., Harris D., Churcher C., Harris D., Harris D., et al. Massive gene decay in the leprosy bacillus. Nature. 2001;409:1007–1011. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous
