Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei
- PMID: 17210949
- PMCID: PMC2063932
- DOI: 10.1083/jcb.200607174
Nuclear repositioning of the VSG promoter during developmental silencing in Trypanosoma brucei
Abstract
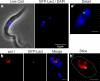
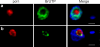
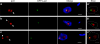
Interphase nuclear repositioning of chromosomes has been implicated in the epigenetic regulation of RNA polymerase (pol) II transcription. However, little is known about the nuclear position-dependent regulation of RNA pol I-transcribed loci. Trypanosoma brucei is an excellent model system to address this question because its two main surface protein genes, procyclin and variant surface glycoprotein (VSG), are transcribed by pol I and undergo distinct transcriptional activation or downregulation events during developmental differentiation. Although the monoallelically expressed VSG locus is exclusively localized to an extranucleolar body in the bloodstream form, in this study, we report that nonmutually exclusive procyclin genes are located at the nucleolar periphery. Interestingly, ribosomal DNA loci and pol I transcription activity are restricted to similar perinucleolar positions. Upon developmental transcriptional downregulation, however, the active VSG promoter selectively undergoes a rapid and dramatic repositioning to the nuclear envelope. Subsequently, the VSG promoter region was subjected to chromatin condensation. We propose a model whereby the VSG expression site pol I promoter is selectively targeted by temporal nuclear repositioning during developmental silencing.
Figures

Similar articles
-
RNA polymerase I transcribes procyclin genes and variant surface glycoprotein gene expression sites in Trypanosoma brucei.Eukaryot Cell. 2003 Jun;2(3):542-51. doi: 10.1128/EC.2.3.542-551.2003. Eukaryot Cell. 2003. PMID: 12796299 Free PMC article.
-
Nuclear architecture underlying gene expression in Trypanosoma brucei.Trends Microbiol. 2007 Jun;15(6):263-70. doi: 10.1016/j.tim.2007.04.004. Epub 2007 May 4. Trends Microbiol. 2007. PMID: 17481901 Review.
-
Bloodstream form-specific up-regulation of silent vsg expression sites and procyclin in Trypanosoma brucei after inhibition of DNA synthesis or DNA damage.J Biol Chem. 2004 Apr 2;279(14):13363-74. doi: 10.1074/jbc.M312307200. Epub 2004 Jan 15. J Biol Chem. 2004. PMID: 14726511
-
Alpha-amanitin-resistant transcription units in trypanosomes: a comparison of promoter sequences for a VSG gene expression site and for the ribosomal RNA genes.Nucleic Acids Res. 1991 Oct 11;19(19):5153-8. doi: 10.1093/nar/19.19.5153. Nucleic Acids Res. 1991. PMID: 1923801 Free PMC article.
-
Transcription of protein-coding genes in trypanosomes by RNA polymerase I.Annu Rev Microbiol. 1997;51:463-89. doi: 10.1146/annurev.micro.51.1.463. Annu Rev Microbiol. 1997. PMID: 9343357 Review.
Cited by
-
Nicotinamide inhibits the lysosomal cathepsin b-like protease and kills African trypanosomes.J Biol Chem. 2013 Apr 12;288(15):10548-57. doi: 10.1074/jbc.M112.449207. Epub 2013 Feb 26. J Biol Chem. 2013. PMID: 23443665 Free PMC article.
-
Shared Mechanisms for Mutually Exclusive Expression and Antigenic Variation by Protozoan Parasites.Front Cell Dev Biol. 2022 Mar 8;10:852239. doi: 10.3389/fcell.2022.852239. eCollection 2022. Front Cell Dev Biol. 2022. PMID: 35350381 Free PMC article. Review.
-
NUP-1 Is a large coiled-coil nucleoskeletal protein in trypanosomes with lamin-like functions.PLoS Biol. 2012;10(3):e1001287. doi: 10.1371/journal.pbio.1001287. Epub 2012 Mar 27. PLoS Biol. 2012. PMID: 22479148 Free PMC article.
-
Inorganic polyphosphate interacts with nucleolar and glycosomal proteins in trypanosomatids.Mol Microbiol. 2018 Dec;110(6):973-994. doi: 10.1111/mmi.14131. Epub 2018 Oct 18. Mol Microbiol. 2018. PMID: 30230089 Free PMC article.
-
Kinetoplastid kinetochore proteins KKT14-KKT15 are divergent Bub1/BubR1-Bub3 proteins.Open Biol. 2024 Jun;14(6):240025. doi: 10.1098/rsob.240025. Epub 2024 Jun 12. Open Biol. 2024. PMID: 38862021 Free PMC article.
References
-
- Acosta-Serrano, A., R.N. Cole, A. Mehlert, M.G. Lee, M.A. Ferguson, and P.T. Englund. 1999. The procyclin repertoire of Trypanosoma brucei. Identification and structural characterization of the Glu-Pro-rich polypeptides. J. Biol. Chem. 274:29763–29771. - PubMed
-
- Alibu, V.P., L. Storm, S. Haile, C. Clayton, and D. Horn. 2005. A doubly inducible system for RNA interference and rapid RNAi plasmid construction in Trypanosoma brucei. Mol. Biochem. Parasitol. 139:75–82. - PubMed
-
- Barry, J.D., and R. McCulloch. 2001. Antigenic variation in trypanosomes: enhanced phenotypic variation in a eukaryotic parasite. Adv. Parasitol. 49:1–70. - PubMed
-
- Borst, P. 2002. Antigenic variation and allelic exclusion. Cell. 109:5–8. - PubMed

