Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution
- PMID: 17213324
- PMCID: PMC1783406
- DOI: 10.1073/pnas.0606318104
Ancestral genome sizes specify the minimum rate of lateral gene transfer during prokaryote evolution
Abstract
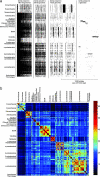
The amount of lateral gene transfer (LGT) that has occurred in microbial evolution is heavily debated. Efforts to quantify LGT through gene-tree comparisons have delivered estimates that between 2% and 60% of all prokaryotic genes have been affected by LGT, the 30-fold discrepancy reflecting differences among gene samples studied and uncertainties inherent in phylogenetic reconstruction. Here we present a simple method that is independent of gene-tree comparisons to estimate the LGT rate among sequenced prokaryotic genomes. If little or no LGT has occurred during evolution, ancestral genome sizes would become unrealistically large, whereas too much LGT would render them far too small. We determine the amount of LGT that is necessary and sufficient to bring the distribution of inferred ancestral genome sizes into agreement with that observed among modern microbes. Rather than testing for phylogenetic congruence or lack thereof across genes, we assume that all gene trees are compatible; hence, our method delivers very conservative lower-bound estimates of the average LGT rate. The results indicate that among 57,670 gene families distributed across 190 sequenced genomes, at least two-thirds and probably all, have been affected by LGT at some time in their evolutionary past. A component of common ancestry nonetheless remains detectable in gene distribution patterns. We estimate the minimum lower bound for the average LGT rate across all genes as 1.1 LGT events per gene family and gene family lifespan and this minimum rate increases sharply when genes present in only a few genomes are excluded from the analysis.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
-
- Ciccarelli FD, Doerks T, von Mering C, Creevey CJ, Snel B, Bork P. Science. 2006;311:1283–1287. - PubMed
-
- Doolittle WF. In: Microbial Phylogeny and Evolution: Concepts and Controversies. Sapp J, editor. New York: Oxford Univ Press; 2004. pp. 119–133.
-
- Gogarten JP, Doolittle WF, Lawrence JG. Mol Biol Evol. 2002;19:2226–2238. - PubMed
Publication types
MeSH terms
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Miscellaneous

