Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles
- PMID: 17214507
- PMCID: PMC1764438
- DOI: 10.1371/journal.pbio.0050008
Large-scale mapping and validation of Escherichia coli transcriptional regulation from a compendium of expression profiles
Abstract
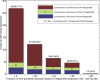
Machine learning approaches offer the potential to systematically identify transcriptional regulatory interactions from a compendium of microarray expression profiles. However, experimental validation of the performance of these methods at the genome scale has remained elusive. Here we assess the global performance of four existing classes of inference algorithms using 445 Escherichia coli Affymetrix arrays and 3,216 known E. coli regulatory interactions from RegulonDB. We also developed and applied the context likelihood of relatedness (CLR) algorithm, a novel extension of the relevance networks class of algorithms. CLR demonstrates an average precision gain of 36% relative to the next-best performing algorithm. At a 60% true positive rate, CLR identifies 1,079 regulatory interactions, of which 338 were in the previously known network and 741 were novel predictions. We tested the predicted interactions for three transcription factors with chromatin immunoprecipitation, confirming 21 novel interactions and verifying our RegulonDB-based performance estimates. CLR also identified a regulatory link providing central metabolic control of iron transport, which we confirmed with real-time quantitative PCR. The compendium of expression data compiled in this study, coupled with RegulonDB, provides a valuable model system for further improvement of network inference algorithms using experimental data.
Conflict of interest statement
Competing interests. A portion of this work was conducted in collaboration with Cellicon Biotechnologies. JJC and TSG are founders and shareholders in the company. GC and JW are also shareholders in the company. All data, results, and algorithms from this collaboration have been made publicly available.
Figures

References
-
- Aderem A. Systems biology: Its practice and challenges. Cell. 2005;121:511–513. - PubMed
-
- Basso K, Margolin AA, Stolovitzky G, Klein U, Dalla-Favera R, et al. Reverse engineering of regulatory networks in human B cells. Nat Genet. 2005;37:382–390. - PubMed
-
- Beer MA, Tavazoie S. Predicting gene expression from sequence. Cell. 2004;117:185–198. - PubMed
-
- de la Fuente A, Brazhnik P, Mendes P. Linking the genes: Inferring quantitative gene networks from microarray data. Trends Genet. 2002;18:395–398. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

