Phylogenetic distribution of translational GTPases in bacteria
- PMID: 17214893
- PMCID: PMC1780047
- DOI: 10.1186/1471-2164-8-15
Phylogenetic distribution of translational GTPases in bacteria
Abstract
Background: Translational GTPases are a family of proteins in which GTPase activity is stimulated by the large ribosomal subunit. Conserved sequence features allow members of this family to be identified.
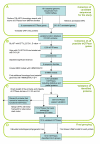
Results: To achieve accurate protein identification and grouping we have developed a method combining searches with Hidden Markov Model profiles and tree based grouping. We found all the genes for translational GTPases in 191 fully sequenced bacterial genomes. The protein sequences were grouped into nine subfamilies. Analysis of the results shows that three translational GTPases, the translation factors EF-Tu, EF-G and IF2, are present in all organisms examined. In addition, several copies of the genes encoding EF-Tu and EF-G are present in some genomes. In the case of multiple genes for EF-Tu, the gene copies are nearly identical; in the case of multiple EF-G genes, the gene copies have been considerably diverged. The fourth translational GTPase, LepA, the function of which is currently unknown, is also nearly universally conserved in bacteria, being absent from only one organism out of the 191 analyzed. The translation regulator, TypA, is also present in most of the organisms examined, being absent only from bacteria with small genomes.Surprisingly, some of the well studied translational GTPases are present only in a very small number of bacteria. The translation termination factor RF3 is absent from many groups of bacteria with both small and large genomes. The specialized translation factor for selenocysteine incorporation--SelB--was found in only 39 organisms. Similarly, the tetracycline resistance proteins (Tet) are present only in a small number of species. Proteins of the CysN/NodQ subfamily have acquired functions in sulfur metabolism and production of signaling molecules. The genes coding for CysN/NodQ proteins were found in 74 genomes. This protein subfamily is not confined to Proteobacteria, as suggested previously but present also in many other groups of bacteria.
Conclusion: Four of the translational GTPase subfamilies (IF2, EF-Tu, EF-G and LepA) are represented by at least one member in each bacterium studied, with one exception in LepA. This defines the set of translational GTPases essential for basic cell functions.
Figures






References
-
- Sergiev PV, Bogdanov AA, Dontsova OA. How can elongation factors EF-G and EF-Tu discriminate the functional state of the ribosome using the same binding site? FEBS Lett. 2005;579:5439–5442. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

