A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection
- PMID: 17215375
- PMCID: PMC1783426
- DOI: 10.1073/pnas.0609954104
A functional SNP of interferon-gamma gene is important for interferon-alpha-induced and spontaneous recovery from hepatitis C virus infection
Erratum in
- Proc Natl Acad Sci U S A. 2007 Mar 13;104(11):4770
Abstract
Cytokine polymorphisms are associated with disease outcome and interferon (IFN) treatment response in hepatitis C virus (HCV) infection. We genotyped eight SNPs spanning the entire IFN-gamma gene in two cohorts and assessed the association between those polymorphisms and treatment response or spontaneous viral clearance. The first cohort was composed of 284 chronically HCV-infected patients who had received IFN-alpha-based therapy and the second was 251 i.v. drug users who had either spontaneously cleared HCV or become chronically infected. A SNP variant located in the proximal IFN-gamma promoter region next to the binding motif of heat shock transcription factor (HSF), -764G, was significantly associated with sustained virological response [P = 0.01, odds ratio (OR) = 2.66 [corrected] (confidence interval 1.3-5.6)[corrected]]. The association was independently significant in multiple logistic regression (P = 0.04) along with race, viral titer, and genotype. This variant was also significantly associated with spontaneous recovery [P = 0.04, OR = 3.51 (1.0-12.5)] in the second cohort. Functional analyses show that the G allele confers a two- to three-fold higher promoter activity and stronger binding affinity to HSF1 than the C allele. Our study suggests that the IFN-gamma promoter SNP -764G/C is functionally important in determining viral clearance and treatment response in HCV-infected patients and may be used as a genetic marker to predict sustained virological response in HCV-infected patients.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
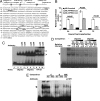
References
-
- Liang TJ, Rehermann B, Seeff LB, Hoofnagle JH. Ann Intern Med. 2000;132:296–305. - PubMed
-
- Tan SL, He Y, Huang Y, Gale M., Jr Curr Opin Pharmacol. 2004;4:465–470. - PubMed
-
- Feld JJ, Hoofnagle JH. Nature. 2005;436:967–972. - PubMed
-
- Promrat K, Liang TJ. Hepatology. 2003;38:1359–1362. - PubMed
-
- Gao B, Hong F, Radaeva S. Hepatology. 2004;39:880–890. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

