Aurora-B regulates RNA methyltransferase NSUN2
- PMID: 17215513
- PMCID: PMC1805108
- DOI: 10.1091/mbc.e06-11-1021
Aurora-B regulates RNA methyltransferase NSUN2
Abstract
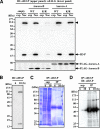
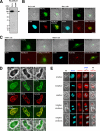
Disassembly of the nucleolus during mitosis is driven by phosphorylation of nucleolar proteins. RNA processing stops until completion of nucleolar reformation in G(1) phase. Here, we describe the RNA methyltransferase NSUN2, a novel substrate of Aurora-B that contains an NOL1/NOP2/sun domain. NSUN2 was concentrated in the nucleolus during interphase and was distributed in the perichromosome and cytoplasm during mitosis. Aurora-B phosphorylated NSUN2 at Ser139. Nucleolar proteins NPM1/nucleophosmin/B23 and nucleolin/C23 were associated with NSUN2 during interphase. In mitotic cells, association between NPM1 and NSUN2 was inhibited, but NSUN2-S139A was constitutively associated with NPM1. The Aurora inhibitor Hesperadin induced association of NSUN2 with NPM1 even in mitosis, despite the silver staining nucleolar organizer region disassembly. In vitro methylation experiments revealed that the Aurora-B-phosphorylation and the phosphorylation-mimic mutation (S139E) suppressed methyltransferase activities of NSUN2. These results indicate that Aurora-B participates to regulate the assembly of nucleolar RNA-processing machinery and the RNA methyltransferase activity of NSUN2 via phosphorylation at Ser139 during mitosis.
Figures






Similar articles
-
The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer.Cells. 2020 Jul 22;9(8):1758. doi: 10.3390/cells9081758. Cells. 2020. PMID: 32708015 Free PMC article. Review.
-
Phosphorylation of multifunctional nucleolar protein nucleophosmin (NPM1) by aurora kinase B is critical for mitotic progression.FEBS Lett. 2014 Jun 27;588(14):2198-205. doi: 10.1016/j.febslet.2014.05.014. Epub 2014 May 21. FEBS Lett. 2014. PMID: 24857377
-
Inactivation of Rho GTPases with Clostridium difficile toxin B impairs centrosomal activation of Aurora-A in G2/M transition of HeLa cells.Mol Biol Cell. 2007 Oct;18(10):3752-63. doi: 10.1091/mbc.e07-03-0281. Epub 2007 Jul 18. Mol Biol Cell. 2007. PMID: 17634283 Free PMC article.
-
Aurora B kinase regulates the postmitotic endoreduplication checkpoint via phosphorylation of the retinoblastoma protein at serine 780.Mol Biol Cell. 2009 Apr;20(8):2218-28. doi: 10.1091/mbc.e08-08-0885. Epub 2009 Feb 18. Mol Biol Cell. 2009. PMID: 19225156 Free PMC article.
-
Functions of the histone chaperone nucleolin in diseases.Subcell Biochem. 2007;41:125-44. doi: 10.1007/1-4020-5466-1_7. Subcell Biochem. 2007. PMID: 17484127 Review.
Cited by
-
The RNA Methyltransferase NSUN2 and Its Potential Roles in Cancer.Cells. 2020 Jul 22;9(8):1758. doi: 10.3390/cells9081758. Cells. 2020. PMID: 32708015 Free PMC article. Review.
-
The transcription factor YY1 is a novel substrate for Aurora B kinase at G2/M transition of the cell cycle.PLoS One. 2012;7(11):e50645. doi: 10.1371/journal.pone.0050645. Epub 2012 Nov 30. PLoS One. 2012. PMID: 23226345 Free PMC article.
-
Landmarks in the Evolution of (t)-RNAs from the Origin of Life up to Their Present Role in Human Cognition.Life (Basel). 2015 Dec 23;6(1):1. doi: 10.3390/life6010001. Life (Basel). 2015. PMID: 26703740 Free PMC article. Review.
-
The Nucleolus Takes Control of Protein Trafficking Under Cellular Stress.Mol Cell Pharmacol. 2010;2(5):203-212. Mol Cell Pharmacol. 2010. PMID: 21499571 Free PMC article.
-
Identification of direct targets and modified bases of RNA cytosine methyltransferases.Nat Biotechnol. 2013 May;31(5):458-64. doi: 10.1038/nbt.2566. Epub 2013 Apr 21. Nat Biotechnol. 2013. PMID: 23604283 Free PMC article.
References
-
- Alexandrov A., Chernyakov I., Gu W., Hiley S. L., Hughes T. R., Grayhack E. J., Phizicky E. M. Rapid tRNA decay can result from lack of nonessential modifications. Mol. Cell. 2006;21:87–96. - PubMed
-
- Andrews P. D., Knatko E., Moore W. J., Swedlow J. R. Mitotic mechanics: the auroras come into view. Curr. Opin. Cell Biol. 2003;15:672–683. - PubMed
-
- Carmena M., Earnshaw W. C. The cellular geography of aurora kinases. Nat. Rev. Mol. Cell Biol. 2003;4:842–854. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

