Histone arginine methylation regulates pluripotency in the early mouse embryo
- PMID: 17215844
- PMCID: PMC3353120
- DOI: 10.1038/nature05458
Histone arginine methylation regulates pluripotency in the early mouse embryo
Abstract
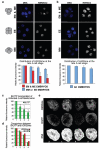
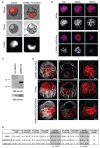
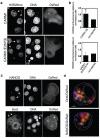
It has been generally accepted that the mammalian embryo starts its development with all cells identical, and only when inside and outside cells form do differences between cells first emerge. However, recent findings show that cells in the mouse embryo can differ in their developmental fate and potency as early as the four-cell stage. These differences depend on the orientation and order of the cleavage divisions that generated them. Because epigenetic marks are suggested to be involved in sustaining pluripotency, we considered that such developmental properties might be achieved through epigenetic mechanisms. Here we show that modification of histone H3, through the methylation of specific arginine residues, is correlated with cell fate and potency. Levels of H3 methylation at specific arginine residues are maximal in four-cell blastomeres that will contribute to the inner cell mass (ICM) and polar trophectoderm and undertake full development when combined together in chimaeras. Arginine methylation of H3 is minimal in cells whose progeny contributes more to the mural trophectoderm and that show compromised development when combined in chimaeras. This suggests that higher levels of H3 arginine methylation predispose blastomeres to contribute to the pluripotent cells of the ICM. We confirm this prediction by overexpressing the H3-specific arginine methyltransferase CARM1 in individual blastomeres and show that this directs their progeny to the ICM and results in a dramatic upregulation of Nanog and Sox2. Thus, our results identify specific histone modifications as the earliest known epigenetic marker contributing to development of ICM and show that manipulation of epigenetic information influences cell fate determination.
Figures

Comment in
-
Developmental biology: marked from the start.Nature. 2007 Jan 11;445(7124):157. doi: 10.1038/445157a. Nature. 2007. PMID: 17215832 No abstract available.
-
Early mouse embryo development: could epigenetics influence cell fate determination?Bioessays. 2007 Jun;29(6):520-4. doi: 10.1002/bies.20591. Bioessays. 2007. PMID: 17508391
References
-
- Piotrowska K, Wianny F, Pedersen RA, Zernicka-Goetz M. Blastomeres arising from the first cleavage division have distinguishable fates in normal mouse development. Development. 2001;128:3739–48. - PubMed
-
- Piotrowska-Nitsche K, Perea-Gomez A, Haraguchi S, Zernicka-Goetz M. Four-cell stage mouse blastomeres have different developmental properties. Development. 2005;132:479–90. - PubMed
-
- Gardner RL. Specification of embryonic axes begins before cleavage in normal mouse development. Development. 2001;128:839–47. - PubMed
-
- Fujimori T, Kurotaki Y, Miyazaki J, Nabeshima Y. Analysis of cell lineage in two- and four-cell mouse embryos. Development. 2003;130:5113–22. - PubMed
-
- Piotrowska-Nitsche K, Zernicka-Goetz M. Spatial arrangement of individual 4-cell stage blastomeres and the order in which they are generated correlate with blastocyst pattern in the mouse embryo. Mech Dev. 2005;122:487–500. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

