Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly
- PMID: 17220203
- PMCID: PMC1820956
- DOI: 10.1105/tpc.106.042739
Chlamydomonas reinhardtii has multiple prolyl 4-hydroxylases, one of which is essential for proper cell wall assembly
Abstract
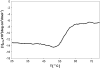
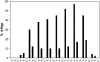
Prolyl 4-hydroxylases (P4Hs) catalyze formation of 4-hydroxyproline (4Hyp), which is found in many plant glycoproteins. We cloned and characterized Cr-P4H-1, one of 10 P4H-like Chlamydomonas reinhardtii polypeptides. Recombinant Cr-P4H-1 is a soluble 29-kD monomer that effectively hydroxylated in vitro both poly(l-Pro) and synthetic peptides representing Pro-rich motifs found in the Chlamydomonas cell wall Hyp-rich glycoprotein (HRGP) GP1. Similar Pro-rich repeats that are likely to be Cr-P4H-1 substrates are also present in the cell wall HRGP GP2 and probably GP3. Suppression of the gene encoding Cr-P4H-1 by RNA interference led to a defective cell wall consisting of a loose network of fibrils resembling the inner and outer W1 and W7 layers of the wild-type wall, while the layers forming the dense central triplet were absent. The lack of Cr-P4H-1 most probably affected 4Hyp content of the major HRPGs of the central triplet, GP1, GP2, and GP3. The reduced 4Hyp levels in these HRGPs can also be expected to affect their glycosylation and, thus, the interactive properties and stabilities of their fibrous shafts. Interestingly, our RNA interference data indicate that the nine other Chlamydomonas P4H-like polypeptides could not fully compensate for the lack of Cr-P4H-1 activity and are therefore likely to have different substrate specificities and functions.
Figures





References
-
- Abrams, E.W., and Andrew, D.J. (2002). Prolyl 4-hydroxylase α-related proteins in Drosophila melanogaster: Tissue-specific embryonic expression of the 99F8-9 cluster. Mech. Dev. 112 165–171. - PubMed
-
- Adair, W.S., and Snell, W.J. (1990). The Chlamydomonas reinhardtii cell wall: Structure, biochemistry, and molecular biology. In Organization and Assembly of Plant and Animal Extracellular Matrix, W.S. Adair and R.P. Mecham, eds (San Diego, CA: Academic Press), pp. 15–84.
-
- Annunen, P., Autio-Harmainen, H., and Kivirikko, K.I. (1998). The novel type II prolyl 4-hydroxylase is the main enzyme form in chondrocytes and capillary endothelial cells, whereas the type I enzyme predominates in most cells. J. Biol. Chem. 273 5989–5992. - PubMed
-
- Annunen, P., Helaakoski, T., Myllyharju, J., Veijola, J., Pihlajaniemi, T., and Kivirikko, K.I. (1997). Cloning of the human prolyl 4-hydroxylase α subunit isoform α(II) and characterization of the type II enzyme tetramer. The α(I) and α(II) subunits do not form a mixed α(I)α(II)β2 tetramer. J. Biol. Chem. 272 17342–17348. - PubMed
-
- Annunen, P., Koivunen, P., and Kivirikko, K.I. (1999). Cloning of the α subunit of prolyl 4-hydroxylase from Drosophila and expression and characterization of the corresponding enzyme tetramer with some unique properties. J. Biol. Chem. 274 6790–6796. - PubMed
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
- Actions
- Actions
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources

