The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae
- PMID: 17220218
- PMCID: PMC1899372
- DOI: 10.1128/JB.01569-06
The rbmBCDEF gene cluster modulates development of rugose colony morphology and biofilm formation in Vibrio cholerae
Abstract
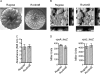
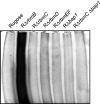
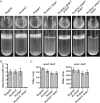
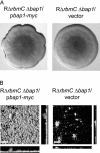
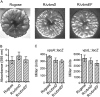
Vibrio cholerae, the causative agent of cholera, can undergo phenotypic variation generating rugose and smooth variants. The rugose variant forms corrugated colonies and well-developed biofilms and exhibits increased levels of resistance to several environmental stresses. Many of these phenotypes are mediated in part by increased expression of the vps genes, which are organized into vps-I and vps-II coding regions, separated by an intergenic region. In this study, we generated in-frame deletions of the five genes located in the vps intergenic region, termed rbmB to -F (rugosity and biofilm structure modulators B to F) in the rugose genetic background, and characterized the mutants for rugose colony development and biofilm formation. Deletion of rbmB, which encodes a protein with low sequence similarity to polysaccharide hydrolases, resulted in an increase in colony corrugation and accumulation of exopolysaccharides relative to the rugose variant. RbmC and its homolog Bap1 are predicted to encode proteins with carbohydrate-binding domains. The colonies of the rbmC bap1 double deletion mutant and bap1 single deletion mutant exhibited a decrease in colony corrugation. Furthermore, the rbmC bap1 double deletion mutant was unable to form biofilms at the air-liquid interface after 2 days, while the biofilms formed on solid surfaces detached readily. Although the colony morphology of rbmDEF mutants was similar to that of the rugose variant, their biofilm structure and cell aggregation phenotypes were different than those of the rugose variant. Taken together, these results indicate that vps intergenic region genes encode proteins that are involved in biofilm matrix production and maintenance of biofilm structure and stability.
Figures
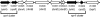





Similar articles
-
Identification and characterization of RbmA, a novel protein required for the development of rugose colony morphology and biofilm structure in Vibrio cholerae.J Bacteriol. 2006 Feb;188(3):1049-59. doi: 10.1128/JB.188.3.1049-1059.2006. J Bacteriol. 2006. PMID: 16428409 Free PMC article.
-
Role of Vibrio polysaccharide (vps) genes in VPS production, biofilm formation and Vibrio cholerae pathogenesis.Microbiology (Reading). 2010 Sep;156(Pt 9):2757-2769. doi: 10.1099/mic.0.040196-0. Epub 2010 May 13. Microbiology (Reading). 2010. PMID: 20466768 Free PMC article.
-
Regulation of rugosity and biofilm formation in Vibrio cholerae: comparison of VpsT and VpsR regulons and epistasis analysis of vpsT, vpsR, and hapR.J Bacteriol. 2007 Jan;189(2):388-402. doi: 10.1128/JB.00981-06. Epub 2006 Oct 27. J Bacteriol. 2007. PMID: 17071756 Free PMC article.
-
[Biofilm development and environmental determinants in Vibrio cholerae].Sheng Wu Gong Cheng Xue Bao. 2017 Sep 25;33(9):1533-1546. doi: 10.13345/j.cjb.170052. Sheng Wu Gong Cheng Xue Bao. 2017. PMID: 28956399 Review. Chinese.
-
Living in the matrix: assembly and control of Vibrio cholerae biofilms.Nat Rev Microbiol. 2015 May;13(5):255-68. doi: 10.1038/nrmicro3433. Nat Rev Microbiol. 2015. PMID: 25895940 Free PMC article. Review.
Cited by
-
Draft Genome Sequence of Vibrio (Listonella) anguillarum ATCC 14181.Genome Announc. 2016 Oct 20;4(5):e01185-16. doi: 10.1128/genomeA.01185-16. Genome Announc. 2016. PMID: 27795288 Free PMC article.
-
Biofilms deform soft surfaces and disrupt epithelia.Elife. 2020 Oct 7;9:e56533. doi: 10.7554/eLife.56533. Elife. 2020. PMID: 33025904 Free PMC article.
-
Cyclic di-GMP Positively Regulates DNA Repair in Vibrio cholerae.J Bacteriol. 2018 Jul 10;200(15):e00005-18. doi: 10.1128/JB.00005-18. Print 2018 Aug 1. J Bacteriol. 2018. PMID: 29610212 Free PMC article.
-
Morphogenesis and cell ordering in confined bacterial biofilms.Proc Natl Acad Sci U S A. 2021 Aug 3;118(31):e2107107118. doi: 10.1073/pnas.2107107118. Proc Natl Acad Sci U S A. 2021. PMID: 34330824 Free PMC article.
-
Facultative control of matrix production optimizes competitive fitness in Pseudomonas aeruginosa PA14 biofilm models.Appl Environ Microbiol. 2015 Dec;81(24):8414-26. doi: 10.1128/AEM.02628-15. Epub 2015 Oct 2. Appl Environ Microbiol. 2015. PMID: 26431965 Free PMC article.
References
-
- Allison, D. G., B. Ruiz, C. SanJose, A. Jaspe, and P. Gilbert. 1998. Extracellular products as mediators of the formation and detachment of Pseudomonas fluorescens biofilms. FEMS Microbiol. Lett. 167:179-184. - PubMed
-
- Baneres, J. L., F. Roquet, A. Martin, and J. Parello. 2000. A minimized human integrin α5β1 that retains ligand recognition. J. Biol. Chem. 275:5888-5903. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

