Crystal structures of a bacterial 6-phosphogluconate dehydrogenase reveal aspects of specificity, mechanism and mode of inhibition by analogues of high-energy reaction intermediates
- PMID: 17222187
- PMCID: PMC6927799
- DOI: 10.1111/j.1742-4658.2006.05585.x
Crystal structures of a bacterial 6-phosphogluconate dehydrogenase reveal aspects of specificity, mechanism and mode of inhibition by analogues of high-energy reaction intermediates
Abstract
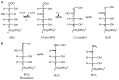
Crystal structures of recombinant Lactococcus lactis 6-phosphogluconate dehydrogenase (LlPDH) in complex with substrate, cofactor, product and inhibitors have been determined. LlPDH shares significant sequence identity with the enzymes from sheep liver and the protozoan parasite Trypanosoma brucei for which structures have been reported. Comparisons indicate that the key residues in the active site are highly conserved, as are the interactions with the cofactor and the product ribulose 5-phosphate. However, there are differences in the conformation of the substrate 6-phosphogluconate which may reflect distinct states relevant to catalysis. Analysis of the complex formed with the potent inhibitor 4-phospho-d-erythronohydroxamic acid, suggests that this molecule does indeed mimic the high-energy intermediate state that it was designed to. The analysis also identified, as a contaminant by-product of the inhibitor synthesis, 4-phospho-d-erythronamide, which binds in similar fashion. LlPDH can now serve as a model system for structure-based inhibitor design targeting the enzyme from Trypanosoma species.
Figures





References
-
- Barrett MP. The pentose phosphate pathway and parasitic protozoa. Parasitol Today. 1997;13:11–16. - PubMed
-
- Hanau S, Rinaldi E, Dallocchio F, Gilbert IH, Dardonville C, Adams MJ, Gover S, Barrett MP. 6-Phosphogluconate dehydrogenase: a target for drugs in African trypanosomes. Curr Med Chem. 2004;11:2639–2650. - PubMed
-
- Gvozdev VA, Gerasimova TI, Kogan GL, Braslavskaya O. Role of the pentose phosphate pathway in metabolism of Drosophila melanogaster elucidated by mutations affecting glucose 6-phosphate and 6-phosphogluconate dehydrogenases. FEBS Lett. 1976;64:85–88. - PubMed
-
- Hughes MB, Lucchesi JC. Genetic rescue of a lethal ‘null’ activity allele of 6-phosphogluconate dehydrogenase in Drosophila melanogaster. Science. 1977;196:1114–1115. - PubMed
-
- Lobo Z, Maitra PK. Pentose phosphate pathway mutants of yeast. Mol Gen Genet. 1982;185:367–368. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

