In silico panning for a non-competitive peptide inhibitor
- PMID: 17222344
- PMCID: PMC1781467
- DOI: 10.1186/1471-2105-8-11
In silico panning for a non-competitive peptide inhibitor
Abstract
Background: Peptide ligands have tremendous therapeutic potential as efficacious drugs. Currently, more than 40 peptides are available in the market for a drug. However, since costly and time-consuming synthesis procedures represent a problem for high-throughput screening, novel procedures to reduce the time and labor involved in screening peptide ligands are required. We propose the novel approach of 'in silico panning' which consists of a two-stage screening, involving affinity selection by docking simulation and evolution of the peptide ligand using genetic algorithms (GAs). In silico panning was successfully applied to the selection of peptide inhibitor for water-soluble quinoprotein glucose dehydrogenase (PQQGDH).
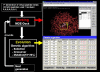
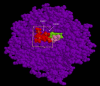
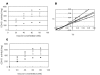
Results: The evolution of peptide ligands for a target enzyme was achieved by combining a docking simulation with evolution of the peptide ligand using genetic algorithms (GAs), which mimic Darwinian evolution. Designation of the target area as next to the substrate-binding site of the enzyme in the docking simulation enabled the selection of a non-competitive inhibitor. In all, four rounds of selection were carried out on the computer; the distribution of the docking energy decreased gradually for each generation and improvements in the docking energy were observed over the four rounds of selection. One of the top three selected peptides with the lowest docking energy, 'SERG' showed an inhibitory effect with Ki value of 20 microM. PQQGDH activity, in terms of the Vmax value, was 3-fold lower than that of the wild-type enzyme in the presence of this peptide. The mechanism of the SERG blockage of the enzyme was identified as non-competitive inhibition. We confirmed the specific binding of the peptide, and its equilibrium dissociation constant (KD) value was calculated as 60 microM by surface plasmon resonance (SPR) analysis.
Conclusion: We demonstrate an effective methodology of in silico panning for the selection of a non-competitive peptide inhibitor from small virtual peptide library. This study is the first to demonstrate the usefulness of in silico evolution using experimental data. Our study highlights the usefulness of this strategy for structure-based screening of enzyme inhibitors.
Figures


Similar articles
-
Peptide ligand screening of alpha-synuclein aggregation modulators by in silico panning.BMC Bioinformatics. 2007 Nov 16;8:451. doi: 10.1186/1471-2105-8-451. BMC Bioinformatics. 2007. PMID: 18005454 Free PMC article.
-
In Silico-Ex Vitro Iteration Strategy for Affinity Maturation of Anti-Ricin Peptides and the SPR Biosensing Application.Toxins (Basel). 2023 Aug 3;15(8):490. doi: 10.3390/toxins15080490. Toxins (Basel). 2023. PMID: 37624247 Free PMC article.
-
Sequential determination of ligands binding to discrete components in heterogeneous mixtures by iterative panning and blocking (IPAB).J Mol Biol. 2000 Feb 25;296(3):821-32. doi: 10.1006/jmbi.1999.3487. J Mol Biol. 2000. PMID: 10677284
-
Combinatorial solid phase peptide synthesis and bioassays.J Biochem Mol Biol. 2005 Sep 30;38(5):517-25. doi: 10.5483/bmbrep.2005.38.5.517. J Biochem Mol Biol. 2005. PMID: 16202229 Review.
-
Phage display biopanning and isolation of target-unrelated peptides: in search of nonspecific binders hidden in a combinatorial library.Amino Acids. 2016 Dec;48(12):2699-2716. doi: 10.1007/s00726-016-2329-6. Epub 2016 Sep 20. Amino Acids. 2016. PMID: 27650972 Review.
Cited by
-
Peptide ligand screening of alpha-synuclein aggregation modulators by in silico panning.BMC Bioinformatics. 2007 Nov 16;8:451. doi: 10.1186/1471-2105-8-451. BMC Bioinformatics. 2007. PMID: 18005454 Free PMC article.
-
Computational Design of Hypothetical New Peptides Based on a Cyclotide Scaffold as HIV gp120 Inhibitor.PLoS One. 2015 Oct 30;10(11):e0139562. doi: 10.1371/journal.pone.0139562. eCollection 2015. PLoS One. 2015. PMID: 26517259 Free PMC article.
-
Microarray Selection of Cooperative Peptides for Modulating Enzyme Activities.Microarrays (Basel). 2017 Apr 26;6(2):8. doi: 10.3390/microarrays6020008. Microarrays (Basel). 2017. PMID: 28445435 Free PMC article.
-
PepComposer: computational design of peptides binding to a given protein surface.Nucleic Acids Res. 2016 Jul 8;44(W1):W522-8. doi: 10.1093/nar/gkw366. Epub 2016 Apr 30. Nucleic Acids Res. 2016. PMID: 27131789 Free PMC article.
-
Computer-aided design of amino acid-based therapeutics: a review.Drug Des Devel Ther. 2018 May 14;12:1239-1254. doi: 10.2147/DDDT.S159767. eCollection 2018. Drug Des Devel Ther. 2018. PMID: 29795978 Free PMC article. Review.
References
-
- V M. WATCHING PEPTIDE DRUGS GROW UP. CHEMICAL& Engineering News. 2005;83:17–24.
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

