Catalytic mechanism of a MYST family histone acetyltransferase
- PMID: 17223684
- PMCID: PMC2752042
- DOI: 10.1021/bi602513x
Catalytic mechanism of a MYST family histone acetyltransferase
Erratum in
- Biochemistry. 2007 Jul 17;46(28):8484
Abstract
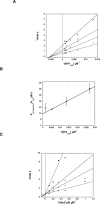
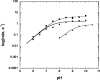
Distinct catalytic mechanisms have been proposed for the Gcn5 and MYST histone acetyltransferase (HAT) families. Gcn5-like HATs utilize an ordered sequential mechanism involving direct nucleophilic attack of the N-epsilon-lysine on the enzyme-bound acetyl-CoA. Recently, MYST enzymes were reported to employ a ping-pong route of catalysis via an acetyl-cysteine intermediate. Here, using the prototypical MYST family member Esa1, and its physiological complex (piccolo NuA4), steady-state kinetic analyses revealed a kinetic mechanism that requires the formation of a ternary complex prior to catalysis, where acetyl-CoA binds first and CoA is the last product released. In the absence of histone acceptor, slow rates of enzyme auto-acetylation (7 x 10(-4) s(-1), or approximately 2500-fold slower than histone acetylation; kcat = 1.6 s(-1)) and of CoA formation (0.0021 s(-1)) were inconsistent with a kinetically competent acetyl-enzyme intermediate. Previously, Cys-304 of Esa1 was the proposed nucleophile that forms an acetyl-cysteine intermediate. Here, mutation of this cysteine (C304A) in Esa1 or within the piccolo NuA4 complex yielded an enzyme that was catalytically indistinguishable from the wild type. Similarly, a pH rate (kcat) analysis of the wild type and C304A revealed an ionization (pKa = 7.6-7.8) that must be unprotonated. Mutation of a conserved active-site glutamate (E338Q) reduced kcat approximately 200-fold at pH 7.5; however, at higher pH, E338Q exhibited nearly wild-type activity. These data are consistent with Glu-338 (general base) activating the N-epsilon-lysine by deprotonation. Together, the results suggest that MYST family HATs utilize a direct-attack mechanism within an Esa1 x acetyl-CoA x histone ternary complex.
Figures

Similar articles
-
Catalysis and substrate selection by histone/protein lysine acetyltransferases.Curr Opin Struct Biol. 2008 Dec;18(6):682-9. doi: 10.1016/j.sbi.2008.11.004. Curr Opin Struct Biol. 2008. PMID: 19056256 Free PMC article. Review.
-
The catalytic mechanism of the ESA1 histone acetyltransferase involves a self-acetylated intermediate.Nat Struct Biol. 2002 Nov;9(11):862-9. doi: 10.1038/nsb849. Nat Struct Biol. 2002. PMID: 12368900
-
Catalytic-site mutations in the MYST family histone Acetyltransferase Esa1.Genetics. 2008 Mar;178(3):1209-20. doi: 10.1534/genetics.107.080135. Epub 2008 Feb 1. Genetics. 2008. PMID: 18245364 Free PMC article.
-
Modulating acetyl-CoA binding in the GCN5 family of histone acetyltransferases.J Biol Chem. 2002 Jul 26;277(30):27337-44. doi: 10.1074/jbc.M203251200. Epub 2002 May 6. J Biol Chem. 2002. PMID: 11994311
-
Catalysis by protein acetyltransferase Gcn5.Biochim Biophys Acta Gene Regul Mech. 2021 Feb;1864(2):194627. doi: 10.1016/j.bbagrm.2020.194627. Epub 2020 Aug 22. Biochim Biophys Acta Gene Regul Mech. 2021. PMID: 32841743 Free PMC article. Review.
Cited by
-
Histone acetyltransferases: Rising ancient counterparts to protein kinases.Biopolymers. 2013 Feb;99(2):98-111. doi: 10.1002/bip.22128. Biopolymers. 2013. PMID: 23175385 Free PMC article. Review.
-
Mechanisms, Detection, and Relevance of Protein Acetylation in Prokaryotes.mBio. 2019 Apr 9;10(2):e02708-18. doi: 10.1128/mBio.02708-18. mBio. 2019. PMID: 30967470 Free PMC article. Review.
-
Metabolic regulation of gene expression through histone acylations.Nat Rev Mol Cell Biol. 2017 Feb;18(2):90-101. doi: 10.1038/nrm.2016.140. Epub 2016 Dec 7. Nat Rev Mol Cell Biol. 2017. PMID: 27924077 Free PMC article. Review.
-
Catalysis and substrate selection by histone/protein lysine acetyltransferases.Curr Opin Struct Biol. 2008 Dec;18(6):682-9. doi: 10.1016/j.sbi.2008.11.004. Curr Opin Struct Biol. 2008. PMID: 19056256 Free PMC article. Review.
-
Enzyme kinetics and inhibition of histone acetyltransferase KAT8.Eur J Med Chem. 2015 Nov 13;105:289-96. doi: 10.1016/j.ejmech.2015.10.016. Epub 2015 Oct 22. Eur J Med Chem. 2015. PMID: 26505788 Free PMC article.
References
-
- Doyon Y, Cote J. The highly conserved and multifunctional NuA4 HAT complex. Curr Opin Genet Dev. 2004;14:147–54. - PubMed
-
- Roth SY, Denu JM, Allis CD. Histone acetyltransferases. Annu Rev Biochem. 2001;70:81–120. - PubMed
-
- Vetting MW, LP S. d. C., Yu M, Hegde SS, Magnet S, Roderick SL, Blanchard JS. Structure and functions of the GNAT superfamily of acetyltransferases. Arch Biochem Biophys. 2005;433:212–26. - PubMed
-
- Marmorstein R. Structure of histone acetyltransferases. J Mol Biol. 2001;311:433–44. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

