Translational control of retroviruses
- PMID: 17224922
- PMCID: PMC7096986
- DOI: 10.1038/nrmicro1599
Translational control of retroviruses
Abstract
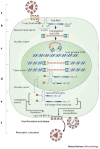
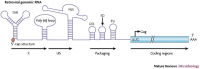
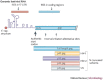
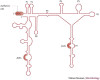
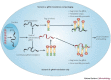
All replication-competent retroviruses contain three main reading frames, gag, pol and env, which are used for the synthesis of structural proteins, enzymes and envelope proteins respectively. Complex retroviruses, such as lentiviruses, also code for regulatory and accessory proteins that have essential roles in viral replication. The concerted expression of these genes ensures the efficient polypeptide production required for the assembly and release of new infectious progeny virions. Retroviral protein synthesis takes place in the cytoplasm and depends exclusively on the translational machinery of the host infected cell. Therefore, not surprisingly, retroviruses have developed RNA structures and strategies to promote robust and efficient expression of viral proteins in a competitive cellular environment.
Conflict of interest statement
The authors declare no competing financial interests.
Figures

References
-
- Telenitsky A, Goff SP. Retroviruses. 1997. pp. 121–160.
-
- Vogt PK. Retroviruses. 1997. pp. 1–25.
-
- Rabson AB, Graves BJ. Retroviruses. 1997. pp. 205–261.
-
- Swanstrom R, Willis JW. Retroviruses. 1997. pp. 263–334.
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

