Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato
- PMID: 17226066
- PMCID: PMC2779359
- DOI: 10.1007/s00705-006-0917-6
Identification and characterisation of tomato torrado virus, a new plant picorna-like virus from tomato
Abstract
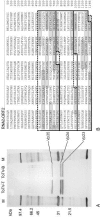
A new virus was isolated from tomato plants from the Murcia region in Spain which showed symptoms of 'torrado disease'; very distinct necrotic, almost burn-like symptoms on leaves of infected plants. The virus particles are isometric with a diameter of approximately 28 nm. The viral genome consists of two (+)ssRNA molecules of 7793 (RNA1) and 5389 nts (RNA2). RNA1 contains one open reading frame (ORF) encoding a predicted polyprotein of 241 kDa that shows conserved regions with motifs typical for a protease-cofactor, a helicase, a protease and an RNA-dependent RNA polymerase. RNA2 contains two, partially overlapping ORFs potentially encoding proteins of 20 and 134 kDa. These viral RNAs are encapsidated by three proteins with estimated sizes of 35, 26 and 23 kDa. Direct protein sequencing mapped these coat proteins to ORF2 on RNA2. Phylogenetic analyses of nucleotide and derived amino acid sequences showed that the virus is related to but distinct from viruses belonging to the genera Sequivirus, Sadwavirus and Cheravirus. This new virus, for which the name tomato torrado virus is proposed, most likely represents a member of a new plant virus genus.
Figures


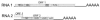
Similar articles
-
Tomato marchitez virus, a new plant picorna-like virus from tomato related to tomato torrado virus.Arch Virol. 2008;153(1):127-34. doi: 10.1007/s00705-007-1076-0. Epub 2007 Oct 29. Arch Virol. 2008. PMID: 17965923
-
Tomato chocolate spot virus, a member of a new torradovirus species that causes a necrosis-associated disease of tomato in Guatemala.Arch Virol. 2010 Jun;155(6):857-69. doi: 10.1007/s00705-010-0653-9. Epub 2010 Apr 9. Arch Virol. 2010. PMID: 20376682 Free PMC article.
-
Identification of the cleavage sites of the RNA2-encoded polyproteins for two members of the genus Torradovirus by N-terminal sequencing of the virion capsid proteins.Virology. 2016 Nov;498:109-115. doi: 10.1016/j.virol.2016.08.014. Epub 2016 Aug 25. Virology. 2016. PMID: 27567259
-
Pepino mosaic virus: a successful pathogen that rapidly evolved from emerging to endemic in tomato crops.Mol Plant Pathol. 2010 Mar;11(2):179-89. doi: 10.1111/j.1364-3703.2009.00600.x. Mol Plant Pathol. 2010. PMID: 20447268 Free PMC article. Review.
-
Impact of coat protein on evolution of ilarviruses.Curr Top Membr. 2024;93:75-84. doi: 10.1016/bs.ctm.2024.05.002. Epub 2024 Jun 4. Curr Top Membr. 2024. PMID: 39181578 Review.
Cited by
-
Enhanced Apiaceous Potyvirus Phylogeny, Novel Viruses, and New Country and Host Records from Sequencing Apiaceae Samples.Plants (Basel). 2022 Jul 27;11(15):1951. doi: 10.3390/plants11151951. Plants (Basel). 2022. PMID: 35956429 Free PMC article.
-
The Torradovirus-specific RNA2-ORF1 protein is necessary for plant systemic infection.Mol Plant Pathol. 2018 Jun;19(6):1319-1331. doi: 10.1111/mpp.12615. Epub 2017 Dec 20. Mol Plant Pathol. 2018. PMID: 28940803 Free PMC article.
-
Genomic Analysis of a Novel Torradovirus "Rehmannia Torradovirus Virus": Two Distinct Variants Infecting Rehmannia glutinosa.Microorganisms. 2024 Aug 11;12(8):1643. doi: 10.3390/microorganisms12081643. Microorganisms. 2024. PMID: 39203485 Free PMC article.
-
Quasispecies nature of Pepino mosaic virus and its evolutionary dynamics.Virus Genes. 2010 Oct;41(2):260-7. doi: 10.1007/s11262-010-0497-0. Epub 2010 Jun 12. Virus Genes. 2010. PMID: 20549323
-
The nucleotide sequence of a Polish isolate of Tomato torrado virus.Virus Genes. 2008 Dec;37(3):400-6. doi: 10.1007/s11262-008-0284-3. Epub 2008 Sep 10. Virus Genes. 2008. PMID: 18781383
References
-
- Fauquet CM, Mayo MA, Maniloff J, Desselberger U, Ball LA (eds) (2005) Virus Taxonomy, Eighth Report of the International Committee on Taxonomy of Viruses. Elsevier Academic Press
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

