Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor
- PMID: 17227865
- PMCID: PMC1783108
- DOI: 10.1073/pnas.0609848104
Histoplasma capsulatum alpha-(1,3)-glucan blocks innate immune recognition by the beta-glucan receptor
Abstract
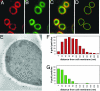
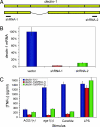
Successful infection by fungal pathogens depends on subversion of host immune mechanisms that detect conserved cell wall components such as beta-glucans. A less common polysaccharide, alpha-(1,3)-glucan, is a cell wall constituent of most fungal respiratory pathogens and has been correlated with pathogenicity or linked directly to virulence. However, the precise mechanism by which alpha-(1,3)-glucan promotes fungal virulence is unknown. Here, we show that alpha-(1,3)-glucan is present in the outermost layer of the Histoplasma capsulatum yeast cell wall and contributes to pathogenesis by concealing immunostimulatory beta-glucans from detection by host phagocytic cells. Production of proinflammatory TNFalpha by phagocytes was suppressed either by the presence of the alpha-(1,3)-glucan layer on yeast cells or by RNA interference based depletion of the host beta-glucan receptor dectin-1. Thus, we have functionally defined key molecular components influencing the initial host-pathogen interaction in histoplasmosis and have revealed an important mechanism by which H. capsulatum thwarts the host immune system. Furthermore, we propose that the degree of this evasion contributes to the difference in pathogenic potential between dimorphic fungal pathogens and opportunistic fungi.
Conflict of interest statement
The authors declare no conflict of interest.
Figures



References
-
- Brown GD. Nat Rev Immunol. 2006;6:33–43. - PubMed
-
- Palma AS, Feizi T, Zhang Y, Stoll MS, Lawson AM, Diaz-Rodriguez E, Campanero-Rhodes MA, Costa J, Gordon S, Brown GD, et al. J Biol Chem. 2006;281:5771–5779. - PubMed
-
- Nemecek JC, Wuthrich M, Klein BS. Science. 2006;312:583–588. - PubMed
-
- Medoff G, Sacco M, Maresca B, Schlessinger D, Painter A, Kobayashi GS, Carratu L. Science. 1986;231:476–479. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

