Involvement of zebrafish Na+,K+ ATPase in myocardial cell junction maintenance
- PMID: 17227894
- PMCID: PMC2063941
- DOI: 10.1083/jcb.200606116
Involvement of zebrafish Na+,K+ ATPase in myocardial cell junction maintenance
Abstract
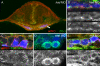
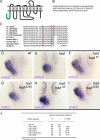
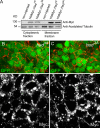
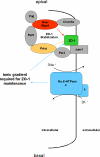
Na(+),K(+) ATPase is an essential ion pump involved in regulating ionic concentrations within epithelial cells. The zebrafish heart and mind (had) mutation, which disrupts the alpha1B1 subunit of Na(+),K(+) ATPase, causes heart tube elongation defects and other developmental abnormalities that are reminiscent of several epithelial cell polarity mutants, including nagie oko (nok). We demonstrate genetic interactions between had and nok in maintaining Zonula occludens-1 (ZO-1)-positive junction belts within myocardial cells. Functional tests and pharmacological inhibition experiments demonstrate that Na(+),K(+) ATPase activity is positively regulated via an N-terminal phosphorylation site that is necessary for correct heart morphogenesis to occur, and that maintenance of ZO-1 junction belts requires ion pump activity. These findings suggest that the correct ionic balance of myocardial cells is essential for the maintenance of epithelial integrity during heart morphogenesis.
Figures


References
-
- Anzenberger, U., N. Bit-Avragim, S. Rohr, F. Rudolph, B. Dehmel, P. Haas, D. Gilmour, T.E. Willnow, and S. Abdelilah-Seyfried. 2006. Elucidation of Megalin/LRP2-dependent endocytic transport processes in the larval zebrafish pronephros. J. Cell Sci. 119:2127–2137. - PubMed
-
- Blanco, G., and R.W. Mercer. 1998. Isozymes of the Na+,K+ ATPase: heterogeneity in structure, diversity in function. Am. J. Physiol. 275:F633–F650. - PubMed
-
- Chen, J., and M. Fishman. 1996. Zebrafish tinman homolog demarcates the heart field and initiates myocardial differentiation. Development. 122:3809–3816. - PubMed
-
- Chibalin, A.V., A.I. Katz, P.O. Berggren, and A.M. Bertorello. 1997. Receptor-mediated inhibition of renal Na+,K+ ATPase is associated with endocytosis of its alpha- and beta-subunits. Am. J. Physiol. 273:C1458–C1465. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

