Effects of base excision repair gene polymorphisms on pancreatic cancer survival
- PMID: 17230526
- PMCID: PMC1892183
- DOI: 10.1002/ijc.22301
Effects of base excision repair gene polymorphisms on pancreatic cancer survival
Abstract
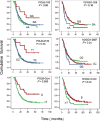
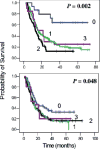
To explore the association between single nucleotide polymorphisms of DNA repair genes and overall survival of patients with pancreatic cancer, we conducted a study in 378 cases of pancreatic adenocarcinoma who were treated at The University of Texas M. D. Anderson Cancer Center between February 1999 and October 2004 and were followed up to April 2006. Genotypes were determined using genomic DNA and the MassCode method. Overall survival was analyzed using the Kaplan-Meier plot, log-rank test and Cox regression. We observed a strong effect of the POLB A165G and T2133C genotypes on overall survival. The median survival time (MST) was 35.7 months for patients carrying at least 1 of the 2 homozygous variant POLB GG or CC genotypes, compared with 14.8 months for those carrying the AA/AG or TT/TC genotypes (p = 0.02, log rank test). The homozygous variants of hOGG1 G2657A, APEX1 D148E and XRCC1 R194W polymorphisms all showed a weak but significant effect on overall survival as demonstrated by either log rank test or multivariate COX regression after adjusting for other potential confounders. In combined genotype analysis, a predominant effect of the POLB homozygous variants on survival was observed. When POLB was not included in the model, a slightly better survival was observed among those carrying none of the adverse genotypes than those carrying at least one of the adverse genotypes. These observations suggest that polymorphisms of base excision repair genes significantly affect the clinical outcome of patients with pancreatic cancer. These observations need to be confirmed in a larger study of homogenous patient population.
(c) 2007 Wiley-Liss, Inc.
Figures
Similar articles
-
Significant effect of homologous recombination DNA repair gene polymorphisms on pancreatic cancer survival.Cancer Res. 2006 Mar 15;66(6):3323-30. doi: 10.1158/0008-5472.CAN-05-3032. Cancer Res. 2006. PMID: 16540687 Free PMC article.
-
Single nucleotide polymorphisms in DNA repair genes XRCC1 and APEX1 in progression and survival of primary cutaneous melanoma patients.Mutat Res. 2009 Feb 10;661(1-2):78-84. doi: 10.1016/j.mrfmmm.2008.11.011. Epub 2008 Nov 27. Mutat Res. 2009. PMID: 19073198
-
Single nucleotide polymorphisms of RecQ1, RAD54L, and ATM genes are associated with reduced survival of pancreatic cancer.J Clin Oncol. 2006 Apr 10;24(11):1720-8. doi: 10.1200/JCO.2005.04.4206. Epub 2006 Mar 6. J Clin Oncol. 2006. PMID: 16520463 Free PMC article. Clinical Trial.
-
Genetic polymorphisms in the base excision repair pathway and cancer risk: a HuGE review.Am J Epidemiol. 2005 Nov 15;162(10):925-42. doi: 10.1093/aje/kwi318. Epub 2005 Oct 12. Am J Epidemiol. 2005. PMID: 16221808 Review.
-
Association of genetic polymorphisms in the base excision repair pathway with lung cancer risk: a meta-analysis.Lung Cancer. 2006 Dec;54(3):267-83. doi: 10.1016/j.lungcan.2006.08.009. Epub 2006 Sep 18. Lung Cancer. 2006. PMID: 16982113 Review.
Cited by
-
Basic principles and technologies for deciphering the genetic map of cancer.World J Surg. 2009 Apr;33(4):615-29. doi: 10.1007/s00268-008-9851-y. World J Surg. 2009. PMID: 19115029 Free PMC article. Review.
-
A replication study and genome-wide scan of single-nucleotide polymorphisms associated with pancreatic cancer risk and overall survival.Clin Cancer Res. 2012 Jul 15;18(14):3942-51. doi: 10.1158/1078-0432.CCR-11-2856. Epub 2012 Jun 4. Clin Cancer Res. 2012. PMID: 22665904 Free PMC article.
-
Is it time to split strategies to treat homologous recombinant deficiency in pancreas cancer?J Gastrointest Oncol. 2016 Oct;7(5):738-749. doi: 10.21037/jgo.2016.05.04. J Gastrointest Oncol. 2016. PMID: 27747088 Free PMC article. Review.
-
An overview of genetic polymorphisms and pancreatic cancer risk in molecular epidemiologic studies.J Epidemiol. 2011;21(1):2-12. doi: 10.2188/jea.je20100090. Epub 2010 Nov 6. J Epidemiol. 2011. PMID: 21071884 Free PMC article. Review.
-
Association between single-nucleotide polymorphisms of OGG1 gene and pancreatic cancer risk in Chinese Han population.Tumour Biol. 2014 Jan;35(1):809-13. doi: 10.1007/s13277-013-1111-6. Epub 2013 Sep 3. Tumour Biol. 2014. Retraction in: Tumour Biol. 2017 Apr 20. doi: 10.1007/s13277-017-5487-6. PMID: 23999824 Retracted.
References
-
- Bardeesy N, DePinho RA. Pancreatic cancer biology and genetics. Nat Rev Cancer. 2002;2:897–909. - PubMed
-
- Phillips KA, Van Bebber SL. Measuring the value of pharmacoge-nomics. Nat Rev Drug Discov. 2005;4:500–9. - PubMed
-
- Bosken CH, Wei Q, Amos CI, Spitz MR. An analysis of DNA repair as a determinant of survival in patients with non small-cell lung cancer. J Natl Cancer Inst. 2002;94:1091–9. - PubMed
-
- Camps C, Sarries C, Roig B, Sanchez JJ, Queralt C, Sancho E, Marti-nez N, Taron M, Rosell R. Assessment of nucleotide excision repair XPD polymorphisms in the peripheral blood of gemcitabine/cisplatin-treated advanced non-small-cell lung cancer patients. Clin Lung Cancer. 2003;4:237–41. - PubMed
-
- Park DJ, Stoehlmacher J, Zhang W, Tsao-Wei DD, Groshen S, Lenz HJ. A xeroderma pigmentosum group D gene polymorphism predicts clinical outcome to platinum-based chemotherapy in patients with advanced colorectal cancer. Cancer Res. 2001;61:8654–8. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Medical
Research Materials
Miscellaneous

