Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production
- PMID: 17233757
- PMCID: PMC1865004
- DOI: 10.1111/j.1600-0854.2006.00524.x
Host ABCE1 is at plasma membrane HIV assembly sites and its dissociation from Gag is linked to subsequent events of virus production
Abstract
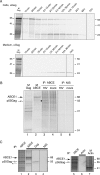
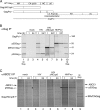
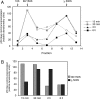
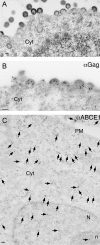
In primate cells, assembly of a single HIV-1 capsid involves multimerization of thousands of Gag polypeptides, typically at the plasma membrane. Although studies support a model in which HIV-1 assembly proceeds through complexes containing Gag and the cellular adenosine triphosphatase ABCE1 (also termed HP68 or ribonuclease L inhibitor), whether these complexes constitute true assembly intermediates remains controversial. Here we demonstrate by pulse labeling in primate cells that a population of Gag associates with endogenous ABCE1 within minutes of translation. In the next approximately 2 h, Gag-ABCE1 complexes increase in size to approximately that of immature capsids. Dissociation of ABCE1 from Gag correlates closely with Gag processing during virion maturation and occurs much less efficiently when the HIV-1 protease is inactivated. Finally, quantitative double-label immunogold electron microscopy reveals that ABCE1 is recruited to sites of assembling wild-type Gag at the plasma membrane but not to sites of an assembly-defective Gag mutant at the plasma membrane. Together these findings demonstrate that a population of Gag present at plasma membrane sites of assembly associates with ABCE1 throughout capsid formation until the onset of virus maturation, which is then followed by virus release. Moreover, the data suggest a linkage between Gag-ABCE1 dissociation and subsequent events of virion production.
Figures




Similar articles
-
Formation of RNA Granule-Derived Capsid Assembly Intermediates Appears To Be Conserved between Human Immunodeficiency Virus Type 1 and the Nonprimate Lentivirus Feline Immunodeficiency Virus.J Virol. 2018 Apr 13;92(9):e01761-17. doi: 10.1128/JVI.01761-17. Print 2018 May 1. J Virol. 2018. PMID: 29467316 Free PMC article.
-
Basic residues in the nucleocapsid domain of Gag are required for interaction of HIV-1 gag with ABCE1 (HP68), a cellular protein important for HIV-1 capsid assembly.J Biol Chem. 2006 Feb 17;281(7):3773-84. doi: 10.1074/jbc.M507255200. Epub 2005 Nov 7. J Biol Chem. 2006. PMID: 16275648
-
Conservation of a stepwise, energy-sensitive pathway involving HP68 for assembly of primate lentivirus capsids in cells.J Virol. 2004 Feb;78(4):1645-56. doi: 10.1128/jvi.78.4.1645-1656.2004. J Virol. 2004. PMID: 14747530 Free PMC article.
-
The biological regulation of ABCE1.IUBMB Life. 2012 Oct;64(10):795-800. doi: 10.1002/iub.1071. IUBMB Life. 2012. PMID: 23008114 Review.
-
Intracellular destinies: degradation, targeting, assembly, and endocytosis of HIV Gag.AIDS Rev. 2007 Jul-Sep;9(3):150-61. AIDS Rev. 2007. PMID: 17982940 Review.
Cited by
-
Budding of retroviruses utilizing divergent L domains requires nucleocapsid.J Virol. 2012 Apr;86(8):4182-93. doi: 10.1128/JVI.07105-11. Epub 2012 Feb 15. J Virol. 2012. PMID: 22345468 Free PMC article.
-
Perturbing HIV-1 Ribosomal Frameshifting Frequency Reveals a cis Preference for Gag-Pol Incorporation into Assembling Virions.J Virol. 2022 Jan 12;96(1):e0134921. doi: 10.1128/JVI.01349-21. Epub 2021 Oct 13. J Virol. 2022. PMID: 34643428 Free PMC article.
-
Sequence requirements for localization and packaging of Ty3 retroelement RNA.Virus Res. 2013 Feb;171(2):319-31. doi: 10.1016/j.virusres.2012.10.008. Epub 2012 Oct 13. Virus Res. 2013. PMID: 23073180 Free PMC article.
-
HIV-1 Gag co-opts a cellular complex containing DDX6, a helicase that facilitates capsid assembly.J Cell Biol. 2012 Aug 6;198(3):439-56. doi: 10.1083/jcb.201111012. Epub 2012 Jul 30. J Cell Biol. 2012. PMID: 22851315 Free PMC article.
-
ABCE1 Regulates RNase L-Induced Autophagy during Viral Infections.Viruses. 2021 Feb 18;13(2):315. doi: 10.3390/v13020315. Viruses. 2021. PMID: 33670646 Free PMC article.
References
-
- Briggs JA, Simon MN, Gross I, Krausslich HG, Fuller SD, Vogt VM, Johnson MC. The stoichiometry of Gag protein in HIV-1. Nat Struct Mol Biol. 2004;11:672–675. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

