Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB
- PMID: 17233885
- PMCID: PMC1784105
- DOI: 10.1186/1471-2199-8-3
Antisense oligonucleotide-induced alternative splicing of the APOB mRNA generates a novel isoform of APOB
Abstract
Background: Apolipoprotein B (APOB) is an integral part of the LDL, VLDL, IDL, Lp(a) and chylomicron lipoprotein particles. The APOB pre-mRNA consists of 29 constitutively-spliced exons. APOB exists as two natural isoforms: the full-length APOB100 isoform, assembled into LDL, VLDL, IDL and Lp(a) and secreted by the liver in humans; and the C-terminally truncated APOB48, assembled into chylomicrons and secreted by the intestine in humans. Down-regulation of APOB100 is a potential therapy to lower circulating LDL and cholesterol levels.
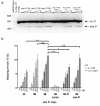
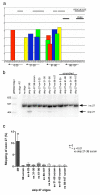
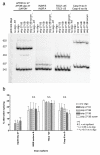
Results: We investigated the ability of 2'O-methyl RNA antisense oligonucleotides (ASOs) to induce the skipping of exon 27 in endogenous APOB mRNA in HepG2 cells. These ASOs are directed towards the 5' and 3' splice-sites of exon 27, the branch-point sequence (BPS) of intron 26-27 and several predicted exonic splicing enhancers within exon 27. ASOs targeting either the 5' or 3' splice-site, in combination with the BPS, are the most effective. The splicing of other alternatively spliced genes are not influenced by these ASOs, suggesting that the effects seen are not due to non-specific changes in alternative splicing. The skip 27 mRNA is translated into a truncated isoform, APOB87SKIP27.
Conclusion: The induction of APOB87SKIP27 expression in vivo should lead to decreased LDL and cholesterol levels, by analogy to patients with hypobetalipoproteinemia. As intestinal APOB mRNA editing and APOB48 expression rely on sequences within exon 26, exon 27 skipping should not affect APOB48 expression unlike other methods of down-regulating APOB100 expression which also down-regulate APOB48.
Figures


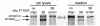
Similar articles
-
Multiple tandem splicing silencer elements suppress aberrant splicing within the long exon 26 of the human Apolipoprotein B gene.BMC Mol Biol. 2013 Feb 7;14:5. doi: 10.1186/1471-2199-14-5. BMC Mol Biol. 2013. PMID: 23391187 Free PMC article.
-
Exon skipping of hepatic APOB pre-mRNA with splice-switching oligonucleotides reduces LDL cholesterol in vivo.Mol Ther. 2013 Mar;21(3):602-9. doi: 10.1038/mt.2012.264. Epub 2013 Jan 15. Mol Ther. 2013. PMID: 23319054 Free PMC article.
-
Studies on the expression of genes encoding apolipoproteins B100 and B48 and the low density lipoprotein receptor in nonhuman primates. Comparison of dietary fat and cholesterol.J Biol Chem. 1989 May 25;264(15):9039-45. J Biol Chem. 1989. PMID: 2722816
-
APOLIPOPROTEIN B: mRNA editing, lipoprotein assembly, and presecretory degradation.Annu Rev Nutr. 2000;20:169-93. doi: 10.1146/annurev.nutr.20.1.169. Annu Rev Nutr. 2000. PMID: 10940331 Review.
-
Homozygous familial hypobetalipoproteinemia: two novel mutations in the splicing sites of apolipoprotein B gene and review of the literature.Atherosclerosis. 2015 Mar;239(1):209-17. doi: 10.1016/j.atherosclerosis.2015.01.014. Epub 2015 Jan 19. Atherosclerosis. 2015. PMID: 25618028 Review.
Cited by
-
Potential therapeutic applications of antisense morpholino oligonucleotides in modulation of splicing in primary immunodeficiency diseases.J Immunol Methods. 2011 Feb 28;365(1-2):1-7. doi: 10.1016/j.jim.2010.12.001. Epub 2010 Dec 13. J Immunol Methods. 2011. PMID: 21147113 Free PMC article. Review.
-
Directing HER4 mRNA expression towards the CYT2 isoform by antisense oligonucleotide decreases growth of breast cancer cells in vitro and in vivo.Br J Cancer. 2013 Jun 11;108(11):2291-8. doi: 10.1038/bjc.2013.247. Epub 2013 May 21. Br J Cancer. 2013. PMID: 23695025 Free PMC article.
-
Targeting Splicing in the Treatment of Human Disease.Genes (Basel). 2017 Feb 24;8(3):87. doi: 10.3390/genes8030087. Genes (Basel). 2017. PMID: 28245575 Free PMC article. Review.
-
CRISPR-SKIP: programmable gene splicing with single base editors.Genome Biol. 2018 Aug 15;19(1):107. doi: 10.1186/s13059-018-1482-5. Genome Biol. 2018. PMID: 30107853 Free PMC article.
-
Alternative Splicing of Nrcam Gene in Dorsal Root Ganglion Contributes to Neuropathic Pain.J Pain. 2020 Jul-Aug;21(7-8):892-904. doi: 10.1016/j.jpain.2019.12.004. Epub 2020 Jan 7. J Pain. 2020. PMID: 31917219 Free PMC article.
References
-
- Rosenson R. Lipoprotein classification; metabolism; and role in atherosclerosis. In: Rose B, editor. UpToDate. Wellesley, MA, USA: UpToDate; 2005.
-
- Rackley C. Pathogenesis of atherosclerosis. In: Rose B, editor. UpToDate. Wellesley, MA, USA: UpToDate; 2005.
-
- Chester A, Scott J, Anant S, Navaratnam N. RNA editing: cytidine to uridine conversion in apolipoprotein B mRNA. Biochim Biophys Acta. 2000;1494:1–13. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous

