The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth
- PMID: 17233889
- PMCID: PMC1794414
- DOI: 10.1186/1471-2180-7-1
The Francisella pathogenicity island protein IglA localizes to the bacterial cytoplasm and is needed for intracellular growth
Abstract
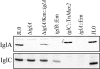
Background: Francisella tularensis is a gram negative, facultative intracellular bacterium that is the etiological agent of tularemia. F. novicida is closely related to F. tularensis but has low virulence for humans while being highly virulent in mice. IglA is a 21 kDa protein encoded by a gene that is part of an iglABCD operon located on the Francisella pathogenicity island (FPI).
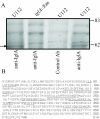
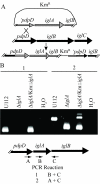
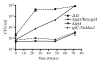
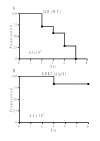
Results: Bioinformatics analysis of the FPI suggests that IglA and IglB are components of a newly described type VI secretion system. In this study, we showed that IglA regulation is controlled by the global regulators MglA and MglB. During intracellular growth IglA production reaches a maximum at about 10 hours post infection. Biochemical fractionation showed that IglA is a soluble cytoplasmic protein and immunoprecipitation experiments demonstrate that it interacts with the downstream-encoded IglB. When the iglB gene was disrupted IglA could not be detected in cell extracts of F. novicida, although IglC could be detected. We further demonstrated that IglA is needed for intracellular growth of F. novicida. A non-polar iglA deletion mutant was defective for growth in mouse macrophage-like cells, and in cis complementation largely restored the wild type macrophage growth phenotype.
Conclusion: The results of this study demonstrate that IglA and IglB are interacting cytoplasmic proteins that are required for intramacrophage growth. The significance of the interaction may be to secrete effector molecules that affect host cell processes.
Figures
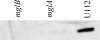



Similar articles
-
Francisella tularensis Live Vaccine Strain deficient in capB and overexpressing the fusion protein of IglA, IglB, and IglC from the bfr promoter induces improved protection against F. tularensis respiratory challenge.Vaccine. 2016 Sep 22;34(41):4969-4978. doi: 10.1016/j.vaccine.2016.08.041. Epub 2016 Aug 28. Vaccine. 2016. PMID: 27577555 Free PMC article.
-
The biochemical properties of the Francisella pathogenicity island (FPI)-encoded proteins IglA, IglB, IglC, PdpB and DotU suggest roles in type VI secretion.Microbiology (Reading). 2011 Dec;157(Pt 12):3483-3491. doi: 10.1099/mic.0.052308-0. Epub 2011 Oct 6. Microbiology (Reading). 2011. PMID: 21980115 Free PMC article.
-
The Francisella tularensis pathogenicity island protein IglC and its regulator MglA are essential for modulating phagosome biogenesis and subsequent bacterial escape into the cytoplasm.Cell Microbiol. 2005 Jul;7(7):969-79. doi: 10.1111/j.1462-5822.2005.00526.x. Cell Microbiol. 2005. PMID: 15953029
-
Molecular and genetic basis of pathogenesis in Francisella tularensis.Ann N Y Acad Sci. 2007 Jun;1105:138-59. doi: 10.1196/annals.1409.010. Epub 2007 Mar 29. Ann N Y Acad Sci. 2007. PMID: 17395737 Review.
-
The Francisella pathogenicity island.Ann N Y Acad Sci. 2007 Jun;1105:122-37. doi: 10.1196/annals.1409.000. Epub 2007 Mar 29. Ann N Y Acad Sci. 2007. PMID: 17395722 Review.
Cited by
-
Atomic structure of T6SS reveals interlaced array essential to function.Cell. 2015 Feb 26;160(5):940-951. doi: 10.1016/j.cell.2015.02.005. Cell. 2015. PMID: 25723168 Free PMC article.
-
Production of outer membrane vesicles and outer membrane tubes by Francisella novicida.J Bacteriol. 2013 Mar;195(6):1120-32. doi: 10.1128/JB.02007-12. Epub 2012 Dec 21. J Bacteriol. 2013. PMID: 23264574 Free PMC article.
-
Subversion of host recognition and defense systems by Francisella spp.Microbiol Mol Biol Rev. 2012 Jun;76(2):383-404. doi: 10.1128/MMBR.05027-11. Microbiol Mol Biol Rev. 2012. PMID: 22688817 Free PMC article. Review.
-
Francisella tularensis T-cell antigen identification using humanized HLA-DR4 transgenic mice.Clin Vaccine Immunol. 2010 Feb;17(2):215-22. doi: 10.1128/CVI.00361-09. Epub 2009 Dec 16. Clin Vaccine Immunol. 2010. PMID: 20016043 Free PMC article.
-
Gene Expression of Type VI Secretion System Associated with Environmental Survival in Acidovorax avenae subsp. avenae by Principle Component Analysis.Int J Mol Sci. 2015 Sep 11;16(9):22008-26. doi: 10.3390/ijms160922008. Int J Mol Sci. 2015. PMID: 26378528 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

