The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export
- PMID: 17234882
- PMCID: PMC1770899
- DOI: 10.1101/gad.1503107
The Spt6 SH2 domain binds Ser2-P RNAPII to direct Iws1-dependent mRNA splicing and export
Abstract
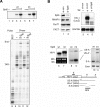
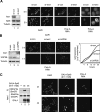
Spt6 promotes transcription elongation at many genes and functions as a histone H3 chaperone to alter chromatin structure during transcription. We show here that mammalian Spt6 binds Ser2-phosphorylated (Ser2P) RNA polymerase II (RNAPII) through a primitive SH2 domain, which recognizes phosphoserine rather than phosphotyrosine residues. Surprisingly, a point mutation in the Spt6 SH2 domain (R1358K) blocked binding to RNAPIIo without affecting transcription elongation rates in vitro. However, HIV-1 and c-myc RNAs formed in cells expressing the mutant Spt6 protein were longer than normal and contained splicing defects. Ectopic expression of the wild-type, but not mutant, Spt6 SH2 domain, caused bulk poly(A)+ RNAs to be retained in the nucleus, further suggesting a widespread role for Spt6 in mRNA processing or assembly of export-competent mRNP particles. We cloned the human Spt6-interacting protein, hIws1 (interacts with Spt6), and found that it associates with the nuclear RNA export factor, REF1/Aly. Depletion of endogenous hIws1 resulted in mRNA processing defects, lower levels of REF1/Aly at the c-myc gene, and nuclear retention of bulk HeLa poly(A)+ RNAs in vivo. Thus binding of Spt6 to Ser2-P RNAPII provides a cotranscriptional mechanism to recruit Iws1, REF1/Aly, and associated mRNA processing, surveillance, and export factors to responsive genes.
Figures





References
-
- Adelman K., Wei W., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T., Wei W., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T., Ardehali M.B., Werner J., Zhu B., Reinberg D., Lis J.T., Werner J., Zhu B., Reinberg D., Lis J.T., Zhu B., Reinberg D., Lis J.T., Reinberg D., Lis J.T., Lis J.T. Drosophila Paf1 modulates chromatin structure at actively transcribed genes. Mol. Cell. Biol. 2006;26:250–260. - PMC - PubMed
-
- Adkins M.W., Tyler J.K., Tyler J.K. Transcriptional activators are dispensable for transcription in the absence of Spt6-mediated chromatin reassembly of promoter regions. Mol. Cell. 2006;21:405–416. - PubMed
-
- Aguilera A. Cotranscriptional mRNP assembly: From the DNA to the nuclear pore. Curr. Opin. Cell Biol. 2005;17:242–250. - PubMed
-
- Ahn S.H., Kim M., Buratowski S., Kim M., Buratowski S., Buratowski S. Phosphorylation of serine 2 within the RNA polymerase II C-terminal domain couples transcription and 3′ end processing. Mol. Cell. 2004;13:67–76. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials
Miscellaneous
