Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide
- PMID: 17237164
- PMCID: PMC1855806
- DOI: 10.1128/JB.01905-06
Functional characterization and membrane topology of Escherichia coli WecA, a sugar-phosphate transferase initiating the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide
Abstract
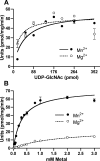
WecA is an integral membrane protein that initiates the biosynthesis of enterobacterial common antigen and O-antigen lipopolysaccharide (LPS) by catalyzing the transfer of N-acetylglucosamine (GlcNAc)-1-phosphate onto undecaprenyl phosphate (Und-P) to form Und-P-P-GlcNAc. WecA belongs to a large family of eukaryotic and prokaryotic prenyl sugar transferases. Conserved aspartic acids in putative cytoplasmic loops 2 (Asp90 and Asp91) and 3 (Asp156 and Asp159) were targeted for replacement mutagenesis with either glutamic acid or asparagine. We examined the ability of each mutant protein to complement O-antigen LPS synthesis in a wecA-deficient strain and also determined the steady-state kinetic parameters of the mutant proteins in an in vitro transfer assay. Apparent K(m) and V(max) values for UDP-GlcNAc, Mg(2+), and Mn(2+) suggest that Asp156 is required for catalysis, while Asp91 appears to interact preferentially with Mg(2+), possibly playing a role in orienting the substrates. Topological analysis using the substituted cysteine accessibility method demonstrated the cytosolic location of Asp90, Asp91, and Asp156 and provided a more refined overall topological map of WecA. Also, we show that cells expressing a WecA derivative C terminally fused with the green fluorescent protein exhibited a punctate distribution of fluorescence on the bacterial surface, suggesting that WecA localizes to discrete regions in the bacterial plasma membrane.
Figures








Similar articles
-
Conserved amino acid residues found in a predicted cytosolic domain of the lipopolysaccharide biosynthetic protein WecA are implicated in the recognition of UDP-N-acetylglucosamine.Microbiology (Reading). 2001 Nov;147(Pt 11):3015-25. doi: 10.1099/00221287-147-11-3015. Microbiology (Reading). 2001. PMID: 11700352
-
Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli.Microbiology (Reading). 2002 Feb;148(Pt 2):571-582. doi: 10.1099/00221287-148-2-571. Microbiology (Reading). 2002. PMID: 11832520
-
Purification and characterization of the bacterial UDP-GlcNAc:undecaprenyl-phosphate GlcNAc-1-phosphate transferase WecA.J Bacteriol. 2008 Nov;190(21):7141-6. doi: 10.1128/JB.00676-08. Epub 2008 Aug 22. J Bacteriol. 2008. PMID: 18723618 Free PMC article.
-
Modeling bacterial UDP-HexNAc: polyprenol-P HexNAc-1-P transferases.Glycobiology. 2005 Sep;15(9):29R-42R. doi: 10.1093/glycob/cwi065. Epub 2005 Apr 20. Glycobiology. 2005. PMID: 15843595 Review.
-
Biosynthesis and Export of Bacterial Glycolipids.Annu Rev Biochem. 2020 Jun 20;89:741-768. doi: 10.1146/annurev-biochem-011520-104707. Annu Rev Biochem. 2020. PMID: 32569526 Review.
Cited by
-
The Mycobacterial Cell Wall--Peptidoglycan and Arabinogalactan.Cold Spring Harb Perspect Med. 2015 Mar 27;5(8):a021113. doi: 10.1101/cshperspect.a021113. Cold Spring Harb Perspect Med. 2015. PMID: 25818664 Free PMC article. Review.
-
Coordination of polymerization, chain termination, and export in assembly of the Escherichia coli lipopolysaccharide O9a antigen in an ATP-binding cassette transporter-dependent pathway.J Biol Chem. 2009 Oct 30;284(44):30662-72. doi: 10.1074/jbc.M109.052878. Epub 2009 Sep 4. J Biol Chem. 2009. PMID: 19734145 Free PMC article.
-
The evolution of antibiotic resistance in an incurable and ultimately fatal infection: A retrospective case study.Evol Med Public Health. 2023 May 6;11(1):163-173. doi: 10.1093/emph/eoad012. eCollection 2023. Evol Med Public Health. 2023. PMID: 37325804 Free PMC article.
-
Natural reversion promotes LPS elongation in an attenuated Coxiella burnetii strain.Nat Commun. 2024 Jan 24;15(1):697. doi: 10.1038/s41467-023-43972-y. Nat Commun. 2024. PMID: 38267444 Free PMC article.
-
Structures and gene clusters of the O-specific polysaccharides of the lipopolysaccharides of Escherichia coli O69 and O146 containing glycolactilic acids: ether conjugates of D-GlcNAc and D-Glc with (R)- and (S)-lactic acid.Glycoconj J. 2017 Feb;34(1):71-84. doi: 10.1007/s10719-016-9730-y. Epub 2016 Sep 19. Glycoconj J. 2017. PMID: 27645300
References
-
- Allingham, J. S., P. A. Pribil, and D. B. Haniford. 1999. All three residues of the Tn10 transposase DDE catalytic triad function in divalent metal ion binding. J. Mol. Biol. 289:1195-1206. - PubMed
-
- Amer, A. O., and M. A. Valvano. 2001. Conserved amino acid residues found in a predicted cytosolic domain of WecA (UDP-N-acetyl glucosamine:undecaprenol-phosphate N-acetylglucosamine-1-phosphate transferase) are implicated in the recognition of UDP-N-acetylglucosamine. Microbiology 147:3015-3025. - PubMed
-
- Amer, A. O., and M. A. Valvano. 2002. Conserved aspartic acids are essential for the enzymic activity of the WecA protein initiating the biosynthesis of O-specific lipopolysaccharide and enterobacterial common antigen in Escherichia coli. Microbiology 148:571-582. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

