The symbiosis regulator CbrA modulates a complex regulatory network affecting the flagellar apparatus and cell envelope proteins
- PMID: 17237174
- PMCID: PMC1855900
- DOI: 10.1128/JB.01834-06
The symbiosis regulator CbrA modulates a complex regulatory network affecting the flagellar apparatus and cell envelope proteins
Abstract
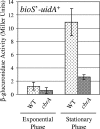
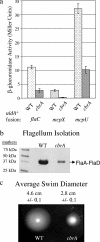
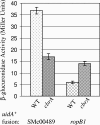
Sinorhizobium meliloti participates in a nitrogen-fixing symbiosis with legume plant host species of the genera Medicago, Melilotus, and Trigonella. We recently identified an S. meliloti two-component sensory histidine kinase, CbrA, which is absolutely required to establish a successful symbiosis with Medicago sativa (K. E. Gibson, G. R. Campbell, J. Lloret, and G. C. Walker, J. Bacteriol. 188:4508-4521, 2006). In addition to having a symbiotic defect, the cbrA::Tn5 mutant also has free-living phenotypes that suggest a cell envelope perturbation. Because the bases for these phenotypes are not well understood, we undertook an identification of CbrA-regulated genes. We performed a microarray analysis and compared the transcriptome of the cbrA::Tn5 mutant to that of the wild type. Our global analysis of gene expression identified 162 genes that are differentially expressed in the cbrA::Tn5 mutant, including those encoding proteins involved in motility and chemotaxis, metabolism, and cell envelope function. With regard to those genes with a known role in symbiosis, we observed increased expression of nine genes with overlapping functions in bacterial invasion of its host, which suggests that the mutant could be competent for invasion. Since these CbrA-repressed genes are vital to the invasion process, it appears that down-regulation of CbrA activity is important at this stage of nodule development. In contrast, our previous work showed that CbrA is required for bacteria to establish themselves within the host as nitrogen-fixing symbionts. Therefore, we propose a model in which CbrA functions as a developmental switch during symbiosis.
Figures


Similar articles
-
CbrA is a stationary-phase regulator of cell surface physiology and legume symbiosis in Sinorhizobium meliloti.J Bacteriol. 2006 Jun;188(12):4508-21. doi: 10.1128/JB.01923-05. J Bacteriol. 2006. PMID: 16740957 Free PMC article.
-
The Sinorhizobium meliloti nitrogen-fixing symbiosis requires CbrA-dependent regulation of a DivL and CckA phosphorelay.J Bacteriol. 2024 Oct 24;206(10):e0039923. doi: 10.1128/jb.00399-23. Epub 2024 Sep 24. J Bacteriol. 2024. PMID: 39315799 Free PMC article.
-
The NtrY/NtrX System of Sinorhizobium meliloti GR4 Regulates Motility, EPS I Production, and Nitrogen Metabolism but Is Dispensable for Symbiotic Nitrogen Fixation.Mol Plant Microbe Interact. 2017 Jul;30(7):566-577. doi: 10.1094/MPMI-01-17-0021-R. Epub 2017 May 17. Mol Plant Microbe Interact. 2017. PMID: 28398840
-
Coordination of symbiosis and cell cycle functions in Sinorhizobium meliloti.Biochim Biophys Acta Gene Regul Mech. 2019 Jul;1862(7):691-696. doi: 10.1016/j.bbagrm.2018.05.003. Epub 2018 May 19. Biochim Biophys Acta Gene Regul Mech. 2019. PMID: 29783033 Review.
-
Sensory transduction to the flagellar motor of Sinorhizobium meliloti.J Mol Microbiol Biotechnol. 2002 May;4(3):183-6. J Mol Microbiol Biotechnol. 2002. PMID: 11931544 Review.
Cited by
-
Coordination of division and development influences complex multicellular behavior in Agrobacterium tumefaciens.PLoS One. 2013;8(2):e56682. doi: 10.1371/journal.pone.0056682. Epub 2013 Feb 20. PLoS One. 2013. PMID: 23437210 Free PMC article.
-
Sinorhizobium meliloti CtrA Stability Is Regulated in a CbrA-Dependent Manner That Is Influenced by CpdR1.J Bacteriol. 2015 Jul;197(13):2139-2149. doi: 10.1128/JB.02593-14. Epub 2015 Apr 20. J Bacteriol. 2015. PMID: 25897034 Free PMC article.
-
A protease and a lipoprotein jointly modulate the conserved ExoR-ExoS-ChvI signaling pathway critical in Sinorhizobium meliloti for symbiosis with legume hosts.PLoS Genet. 2023 Oct 23;19(10):e1010776. doi: 10.1371/journal.pgen.1010776. eCollection 2023 Oct. PLoS Genet. 2023. PMID: 37871041 Free PMC article.
-
Molecular determinants of a symbiotic chronic infection.Annu Rev Genet. 2008;42:413-41. doi: 10.1146/annurev.genet.42.110807.091427. Annu Rev Genet. 2008. PMID: 18983260 Free PMC article. Review.
-
Quantitative proteomic analysis of the Hfq-regulon in Sinorhizobium meliloti 2011.PLoS One. 2012;7(10):e48494. doi: 10.1371/journal.pone.0048494. Epub 2012 Oct 30. PLoS One. 2012. PMID: 23119037 Free PMC article.
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl (ed.). 1991. Current protocols in molecular biology, vol. 1. John Wiley & Sons Inc., Hoboken, NJ.
-
- Barnett, M. J., and R. F. Fisher. 2006. Global gene expression in the rhizobial-legume symbiosis. Symbiosis 42:1-24.
-
- Batut, J., S. G. Andersson, and D. O'Callaghan. 2004. The evolution of chronic infection strategies in the alpha-proteobacteria. Nat. Rev. Microbiol. 2:933-945. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

