The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity
- PMID: 17237350
- PMCID: PMC1820957
- DOI: 10.1105/tpc.106.047720
The Arabidopsis irregular xylem8 mutant is deficient in glucuronoxylan and homogalacturonan, which are essential for secondary cell wall integrity
Abstract
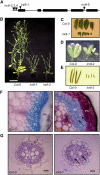
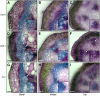
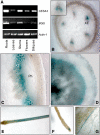
The secondary cell wall in higher plants consists mainly of cellulose, lignin, and xylan and is the major component of biomass in many species. The Arabidopsis thaliana irregular xylem8 (irx8) mutant is dwarfed and has a significant reduction in secondary cell wall thickness. IRX8 belongs to a subgroup of glycosyltransferase family 8 called the GAUT1-related gene family, whose members include GAUT1, a homogalacturonan galacturonosyltransferase, and GAUT12 (IRX8). Here, we use comparative cell wall analyses to show that the irx8 mutant contains significantly reduced levels of xylan and homogalacturonan. Immunohistochemical analyses confirmed that the level of xylan was significantly reduced in the mutant. Structural fingerprinting of the cell wall polymers further revealed that irx8 is deficient in glucuronoxylan. To explore the biological function of IRX8, we crossed irx8 with irx1 (affecting cellulose synthase 8). The homozygous irx1 irx8 exhibited severely dwarfed phenotypes, suggesting that IRX8 is essential for cell wall integrity during cellulose deficiency. Taken together, the data presented show that IRX8 affects the level of glucuronoxylan and homogalacturonan in higher plants and that IRX8 provides an important link between the xylan polymer and the secondary cell wall matrix and directly affects secondary cell wall integrity.
Figures







References
-
- Alonso, J., et al. (2003). Genome-wide insertional mutagenesis of Arabidopsis thaliana. Science 301 653–657. - PubMed
-
- Awano, T., Takabe, K., and Fujita, M. (2002). Xylan deposition on secondary wall of Fagus crenata fiber. Protoplasma 219 106–115. - PubMed
-
- Bauer, S., Vasu, P., Mort, A.J., and Somerville, C.R. (2005). Cloning, expression, and characterization of an oligoxyloglucan reducing end-specific xyloglucanobiohydrolase from Aspergillus nidulans. Carbohydr. Res. 340 2590–2597. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

