Comparison of siRNA-induced off-target RNA and protein effects
- PMID: 17237357
- PMCID: PMC1800510
- DOI: 10.1261/rna.352507
Comparison of siRNA-induced off-target RNA and protein effects
Abstract
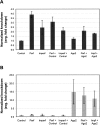
The downregulation of many mRNAs has been observed through bioinformatic analysis of microarray results following transfection of short interfering RNAs (siRNAs). Many of these mRNA changes are due to the interaction of the siRNA guide strand with partially complementary sites and thus are considered "off-target" effects. To examine the mRNA:siRNA interactions important for off-target effects, we generated a panel of mRNA:siRNA combinations containing single and double mismatches, bulges, and noncanonical base-pairing interactions in the 9th, 10th, and 11th positions of two siRNA binding sites located in the 3' UTR of an integrated reporter gene. Approximately half of the mRNA:siRNA combinations containing mismatches in positions 9-11 result in a twofold or more mRNA decrease with varying degrees of protein knockdown. However, mRNA and protein analysis of the various mRNA:siRNA combinations reveals instances in which mRNA and protein levels do not correlate. Analysis of the resulting degradation products recovered from an imperfectly complementary siRNA interaction with an endogenous gene reveals a small fraction of products that map to the canonical siRNA cleavage site. Furthermore, downregulation of ARGONAUTE 2 (AGO2), the only AGO family protein known to catalyze canonical siRNA-mediated cleavage, did not significantly affect the degree of mRNA knockdown observed for one of the stably expressed reporters after transfection of an imperfectly complementary siRNA. These results indicate that although some degree of canonical siRNA cleavage can take place between a siRNA and an off-target transcript, most off-target mRNA reductions are likely attributable to AGO2-independent degradation processes.
Figures


References
-
- Bagga, S., Bracht, J., Hunter, S., Massirer, K., Holtz, J., Eachus, R., Pasquinelli, A.E. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122:553–563. - PubMed
-
- Birmingham, A., Anderson, E.M., Reynolds, A., Ilsley-Tyree, D., Leake, D., Fedorov, Y., Baskerville, S., Maksimova, E., Robinson, K., Karpilow, J., et al. 3′ UTR seed matches, but not overall identity, are associated with RNAi off-targets. Nat. Methods. 2006;3:199–204. - PubMed
-
- Brennecke, J., Hipfner, D.R., Stark, A., Russell, R.B., Cohen, S.M. bantam encodes a developmentally regulated microRNA that controls cell proliferation and regulates the proapoptotic gene hid in Drosophila . Cell. 2003;113:25–36. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
